8WZ9
 
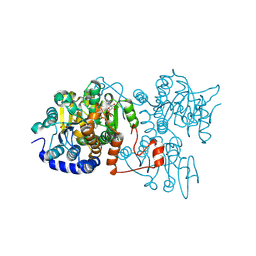 | | The crystal structure of Legionella pneumophila adenosylhomocysteinase Lpg2021 in ternary complex with NAD and adenine | | Descriptor: | ADENINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Gao, Y.S, Xie, R, Chen, Y.N, Ma, J.M, Ge, H.H. | | Deposit date: | 2023-11-01 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | The crystal structure of Legionella pneumophila adenosylhomocysteinase Lpg2021 in ternary complex with NAD and adenine
To Be Published
|
|
8WWG
 
 | |
8WZ8
 
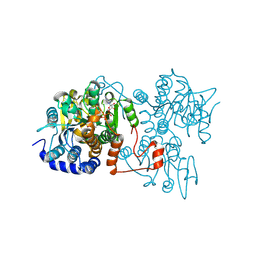 | | The crystal structure of Legionella pneumophila adenosylhomocysteinase Lpg2021(T67A,Q69A) complex with NAD | | Descriptor: | Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Gao, Y.S, Xie, R, Chen, Y.N, Ma, J.M, Ge, H.H. | | Deposit date: | 2023-11-01 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.583 Å) | | Cite: | The crystal structure of Legionella pneumophila adenosylhomocysteinase Lpg2021(T67A,Q69A) complex with NAD
To Be Published
|
|
8WZ6
 
 | | The crystal structure of Legionella pneumophila adenosylhomocysteinase Lpg2021 in ternary complex with NAD and DZNep | | Descriptor: | Adenosylhomocysteinase, DZNep, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Gao, Y.S, Xie, R, Chen, Y.N, Ma, J.M, Ge, H.H. | | Deposit date: | 2023-11-01 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The crystal structure of Legionella pneumophila adenosylhomocysteinase Lpg2021 in ternary complex with NAD and DZNep
To Be Published
|
|
5ZCJ
 
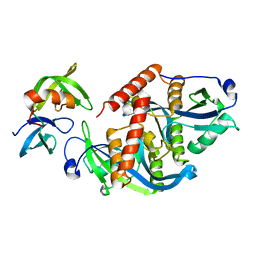 | | Crystal structure of complex | | Descriptor: | TP53-binding protein 1, Tudor-interacting repair regulator protein | | Authors: | Wang, J, Yuan, Z, Cui, Y, Xie, R, Wang, M, Ma, Y, Yu, X, Liu, X. | | Deposit date: | 2018-02-17 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Crystal structure of complex
To Be Published
|
|
5JAU
 
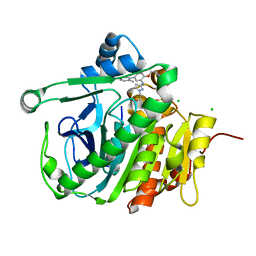 | |
5JAT
 
 | |
5JAN
 
 | |
5JAR
 
 | |
5JAL
 
 | |
5JAS
 
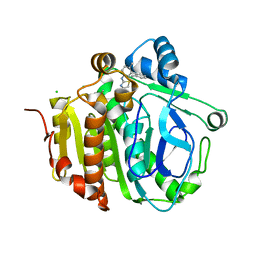 | |
5JAO
 
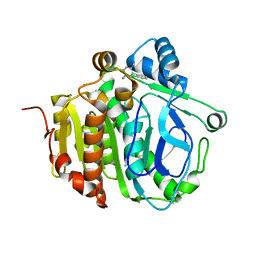 | |
5JAP
 
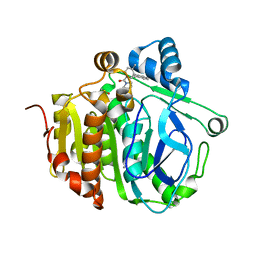 | |
5JAD
 
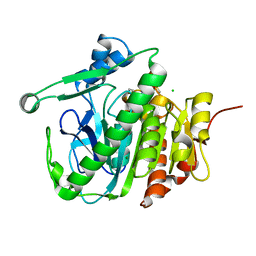 | |
5JAH
 
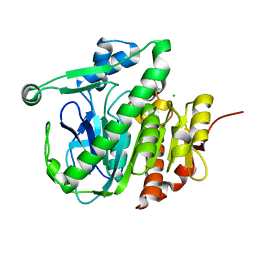 | |
7BQF
 
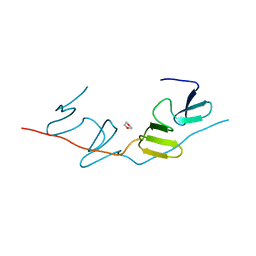 | | Dimerization of SAV1 WW tandem | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Protein salvador homolog 1 | | Authors: | Lin, Z, Zhang, M. | | Deposit date: | 2020-03-24 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.70037615 Å) | | Cite: | A WW Tandem-Mediated Dimerization Mode of SAV1 Essential for Hippo Signaling.
Cell Rep, 32, 2020
|
|
7BQG
 
 | | Complex structure of SAV1 and Dendrin | | Descriptor: | POTASSIUM ION, Protein salvador homolog 1,Dendrin | | Authors: | Lin, Z, Zhang, M. | | Deposit date: | 2020-03-24 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55010867 Å) | | Cite: | A WW Tandem-Mediated Dimerization Mode of SAV1 Essential for Hippo Signaling.
Cell Rep, 32, 2020
|
|
8RFH
 
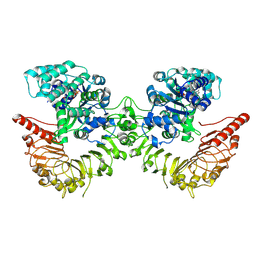 | |
2FTD
 
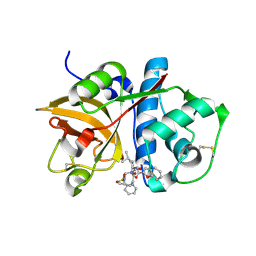 | | Crystal structure of Cathepsin K complexed with 7-Methyl-Substituted Azepan-3-one compound | | Descriptor: | Cathepsin K, N-[(1S)-1-({[(3S,4S,7R)-3-HYDROXY-7-METHYL-1-(PYRIDIN-2-YLSULFONYL)-2,3,4,7-TETRAHYDRO-1H-AZEPIN-4-YL]AMINO}CARBONYL)-3-METHYLBUTYL]-1-BENZOFURAN-2-CARBOXAMIDE | | Authors: | Yamashita, D.S, Baoguang, Z. | | Deposit date: | 2006-01-24 | | Release date: | 2007-01-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure activity relationships of 5-, 6-, and 7-methyl-substituted azepan-3-one cathepsin K inhibitors.
J.Med.Chem., 49, 2006
|
|
5HG1
 
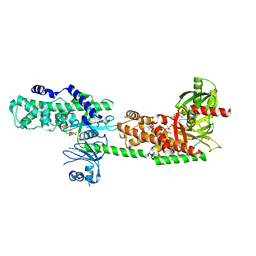 | | Crystal Structure of Human Hexokinase 2 with cmpd 1, a C-2-substituted glucosamine | | Descriptor: | 2-deoxy-2-{[(2E)-3-(3,4-dichlorophenyl)prop-2-enoyl]amino}-alpha-D-glucopyranose, 6-O-phosphono-beta-D-glucopyranose, CITRATE ANION, ... | | Authors: | Campobasso, N, Zhao, B, Smallwood, A. | | Deposit date: | 2016-01-07 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Discovery of a Novel 2,6-Disubstituted Glucosamine Series of Potent and Selective Hexokinase 2 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
5HFU
 
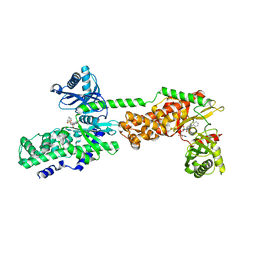 | | Crystal Structure of Human Hexokinase 2 with cmpd 27, a 2-amido-6-benzenesulfonamide glucosamine | | Descriptor: | Hexokinase-2, ~{N}-[(2~{S},3~{R},4~{R},5~{S},6~{R})-6-[[(4-cyanophenyl)sulfonylamino]methyl]-2,4,5-tris(oxidanyl)oxan-3-yl]-3-phenyl-benzamide | | Authors: | Campobasso, N, Zhao, B, Smallwood, A. | | Deposit date: | 2016-01-07 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.923 Å) | | Cite: | Discovery of a Novel 2,6-Disubstituted Glucosamine Series of Potent and Selective Hexokinase 2 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
5HEX
 
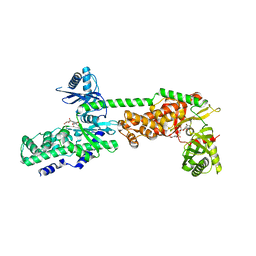 | | Crystal Structure of Human Hexokinase 2 with cmpd 30, a 2-amino-6-benzenesulfonamide glucosamine | | Descriptor: | 2-[(3-bromobenzene-1-carbonyl)amino]-6-{[(4-carboxy-5-methylfuran-2-yl)sulfonyl]amino}-2,6-dideoxy-alpha-D-glucopyranos e, Hexokinase-2 | | Authors: | Campobasso, N, Zhao, B, Smallwood, A. | | Deposit date: | 2016-01-06 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.734 Å) | | Cite: | Discovery of a Novel 2,6-Disubstituted Glucosamine Series of Potent and Selective Hexokinase 2 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
5ZQY
 
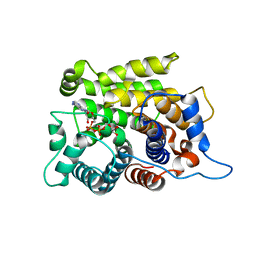 | | Crystal structure of a poly(ADP-ribose) glycohydrolase | | Descriptor: | MAGNESIUM ION, Poly(ADP-ribose) glycohydrolase ARH3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Wang, M, Yuan, Z, Ma, Y, Wang, J, Liu, X. | | Deposit date: | 2018-04-20 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.577 Å) | | Cite: | Structure-function analyses reveal the mechanism of the ARH3-dependent hydrolysis of ADP-ribosylation.
J. Biol. Chem., 293, 2018
|
|
6J68
 
 | | Structure of KIBRA and LATS1 Complex | | Descriptor: | Peptide from Serine/threonine-protein kinase LATS1, Protein KIBRA | | Authors: | Lin, Z, Yang, Z, Ji, Z, Zhang, M. | | Deposit date: | 2019-01-14 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Decoding WW domain tandem-mediated target recognitions in tissue growth and cell polarity.
Elife, 8, 2019
|
|
6JJY
 
 | | Crystal Structure of KIBRA and beta-Dystroglycan | | Descriptor: | Peptide from Dystroglycan, Protein KIBRA, SULFATE ION | | Authors: | Lin, Z, Yang, Z, Ji, Z, Zhang, M. | | Deposit date: | 2019-02-27 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Decoding WW domain tandem-mediated target recognitions in tissue growth and cell polarity.
Elife, 8, 2019
|
|
