1F9X
 
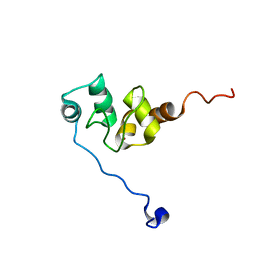 | | AVERAGE NMR SOLUTION STRUCTURE OF THE BIR-3 DOMAIN OF XIAP | | Descriptor: | INHIBITOR OF APOPTOSIS PROTEIN XIAP, ZINC ION | | Authors: | Sun, C, Cai, M, Meadows, R.P, Fesik, S.W. | | Deposit date: | 2000-07-11 | | Release date: | 2001-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure and mutagenesis of the third Bir domain of the inhibitor of apoptosis protein XIAP.
J.Biol.Chem., 275, 2000
|
|
1JHB
 
 | |
1XOX
 
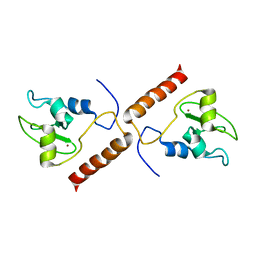 | | SOLUTION STRUCTURE OF HUMAN SURVIVIN | | Descriptor: | Apoptosis inhibitor survivin, ZINC ION | | Authors: | Sun, C, Nettesheim, D, Liu, Z, Olejniczak, E.T. | | Deposit date: | 2004-10-07 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human survivin and its binding interface with smac/diablo
Biochemistry, 44, 2005
|
|
9CVF
 
 | |
9CVG
 
 | |
9CVE
 
 | |
8FOI
 
 | | Native GABA-A receptor from the mouse brain, alpha1-beta2-gamma2 subtype, in complex with GABA and allopregnanolone | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, DODECANE, ... | | Authors: | Sun, C, Gouaux, E. | | Deposit date: | 2022-12-30 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
8G4N
 
 | | Native GABA-A receptor from the mouse brain, alpha1-beta2-gamma2 subtype, in complex with GABA, Zolpidem, and endogenous neurosteroids | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, DODECANE, ... | | Authors: | Sun, C, Gouaux, E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
8G4X
 
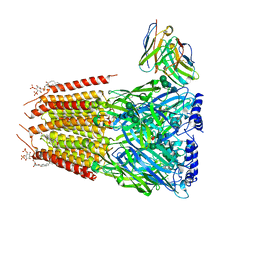 | | Native GABA-A receptor from the mouse brain, meta-alpha1-alpha3-beta2-gamma2 subtype, in complex with GABA and allopregnanolone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, C, Gouaux, E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
8G5F
 
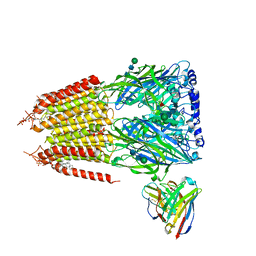 | | Native GABA-A receptor from the mouse brain, ortho-alpha1-alpha3-beta2-gamma2 subtype, in complex with GABA and allopregnanolone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Gamma-aminobutyric acid receptor subunit alpha-1, ... | | Authors: | Sun, C, Gouaux, E. | | Deposit date: | 2023-02-13 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
8G5G
 
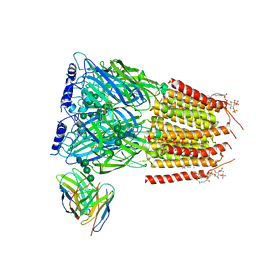 | | Native GABA-A receptor from the mouse brain, meta-alpha1-alpha3-beta2-gamma2 subtype, in complex with GABA, Zolpidem, and endogenous neurosteroid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Gamma-aminobutyric acid receptor subunit alpha-1, ... | | Authors: | Sun, C, Gouaux, E. | | Deposit date: | 2023-02-13 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
8G4O
 
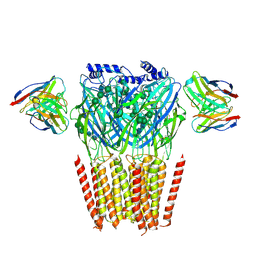 | | Native GABA-A receptor from the mouse brain, alpha1-beta2-gamma2 subtype, in complex with didesethylflurazepam and endogenous GABA | | Descriptor: | (5M)-1-(2-aminoethyl)-7-chloro-5-(2-fluorophenyl)-1,3-dihydro-2H-1,4-benzodiazepin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Sun, C, Gouaux, E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
8G5H
 
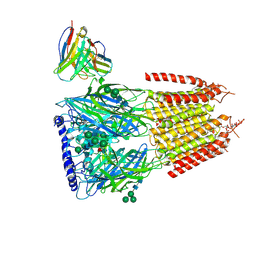 | | Native GABA-A receptor from the mouse brain, ortho-alpha1-alpha3-beta2-gamma2 subtype, in complex with GABA, Zolpidem, and endogenous neurosteroid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Gamma-aminobutyric acid receptor subunit alpha-1, ... | | Authors: | Sun, C, Gouaux, E. | | Deposit date: | 2023-02-13 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
6BTM
 
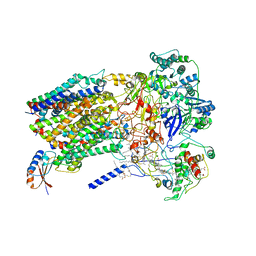 | | Structure of Alternative Complex III from Flavobacterium johnsoniae (Wild Type) | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl ditetradecanoate, Alternative Complex III subunit A, Alternative Complex III subunit B, ... | | Authors: | Sun, C, Benlekbir, S, Venkatakrishnan, P, Yuhang, W, Tajkhorshid, E, Rubinstein, J.L, Gennis, R.B. | | Deposit date: | 2017-12-07 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the alternative complex III in a supercomplex with cytochrome oxidase.
Nature, 557, 2018
|
|
8VJS
 
 | |
8VGR
 
 | |
8VJR
 
 | |
8J2G
 
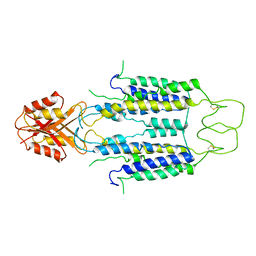 | | Human ZnT1-D47N/D255N mutant | | Descriptor: | Proton-coupled zinc antiporter SLC30A1 | | Authors: | Sun, L, Liu, X, Sun, C, He, B. | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the calcium-coupled zinc export of human ZnT1.
Sci Adv, 10, 2024
|
|
8KFA
 
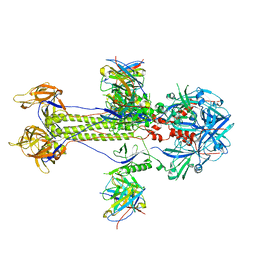 | | Cryo-EM structure of HSV-1 gB with D48 Fab complex | | Descriptor: | D48 heavy chain, D48 light chain, Envelope glycoprotein B | | Authors: | Yang, J, Sun, C, Fang, X, Zeng, M, Liu, Z. | | Deposit date: | 2023-08-15 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | The structure of HSV-1 gB bound to a potent neutralizing antibody reveals a conservative antigenic domain across herpesviruses
hlife, 2023
|
|
6VZA
 
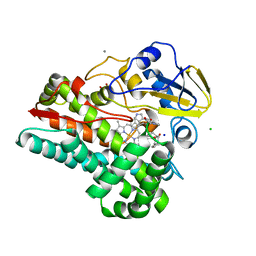 | | Crystal structure of cytochrome P450 NasF5053 Q65I-A86G mutant variant from Streptomyces sp. NRRL F-5053 in the cyclo-L-Trp-L-Pro-bound state | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | Deposit date: | 2020-02-28 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
6VXV
 
 | | Crystal structure of cyclo-L-Trp-L-Pro-bound cytochrome P450 NasF5053 from Streptomyces sp. NRRL F-5053 | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | Deposit date: | 2020-02-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
6W0S
 
 | | Crystal structure of substrate free cytochrome P450 NasF5053 from Streptomyces sp. NRRL F-5053 | | Descriptor: | BROMIDE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | Deposit date: | 2020-03-02 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
6VZB
 
 | | Crystal structure of cytochrome P450 NasF5053 S284A-V288A mutant variant from Streptomyces sp. NRRL F-5053 in the cyclo-L-Trp-L-Pro-bound state | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | Deposit date: | 2020-02-28 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
2JOD
 
 | | Pac1-Rshort N-terminal EC domain Pacap(6-38) complex | | Descriptor: | Pituitary adenylate cyclase-activating polypeptide, Pituitary adenylate cyclase-activating polypeptide type I receptor | | Authors: | Olejniczak, E.T, Sun, C, Song, D, Davis-Taber, R.A, Barrett, L.W, Scott, V.E, Richardson, P.L, Pereda-lopez, A, Uchic, M.E, Solomon, L.R, Lake, M.R, Walter, K.A, Hajduk, P.J. | | Deposit date: | 2007-03-07 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure and mutational analysis of pituitary adenylate cyclase-activating polypeptide binding to the extracellular domain of PAC1-RS.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3NEW
 
 | | p38-alpha complexed with Compound 10 | | Descriptor: | 4-(trifluoromethyl)-3-[3-(trifluoromethyl)phenyl]-1,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, Mitogen-activated protein kinase 14 | | Authors: | Goedken, E.R, Comess, K.M, Sun, C, Argiriadi, M, Jia, Y, Quinn, C.M, Banach, D.L, Marcotte, D, Borhani, D. | | Deposit date: | 2010-06-09 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and Characterization of Non-ATP Site Inhibitors of the Mitogen Activated Protein (MAP) Kinases.
Acs Chem.Biol., 6, 2011
|
|
