6Z1H
 
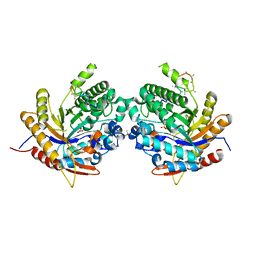 | | Ancestral glycosidase (family 1) | | Descriptor: | ANCESTRAL RECONSTRUCTED GLYCOSIDASE, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Gavira, J.A, Risso, V.A, Sanchez-Ruiz, J.M, Gamiz-Arco, G, Gutierrez-Rus, L, Ibarra-Molero, B, Hoshino, Y, Petrovic, D, Romero-Rivera, A, Seelig, B, Kamerlin, S.C.L, Gaucher, E.A. | | Deposit date: | 2020-05-13 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Heme-binding enables allosteric modulation in an ancient TIM-barrel glycosidase.
Nat Commun, 12, 2021
|
|
7OOL
 
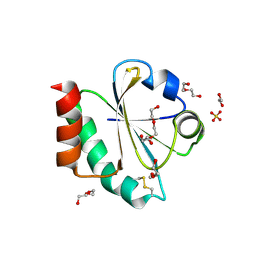 | | Crystal structure of a Candidatus photodesmus katoptron thioredoxin chimera containing an ancestral loop | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Gavira, J.A, Ibarra-Molero, B, Gamiz-Arco, G, Risso, V, Sanchez-Ruiz, J.M. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Combining Ancestral Reconstruction with Folding-Landscape Simulations to Engineer Heterologous Protein Expression.
J.Mol.Biol., 433, 2021
|
|
6Z1M
 
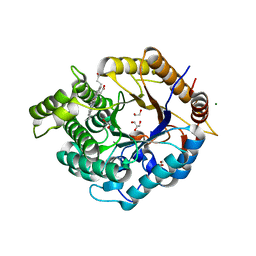 | | Structure of an Ancestral glycosidase (family 1) bound to heme | | Descriptor: | 1,2-ETHANEDIOL, Ancestral reconstructed glycosidase, GLYCEROL, ... | | Authors: | Gavira, J.A, Risso, V.A, Sanchez-Ruiz, J.M, Gamiz-Arco, G, Gutierrez-Rus, L, Ibarra-Molero, B, Oshino, Y, Petrovic, D, Romero-Rivera, A, Seelig, B, Kamerlin, S.C.L, Gaucher, E.A. | | Deposit date: | 2020-05-14 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Heme-binding enables allosteric modulation in an ancient TIM-barrel glycosidase.
Nat Commun, 12, 2021
|
|
4ULX
 
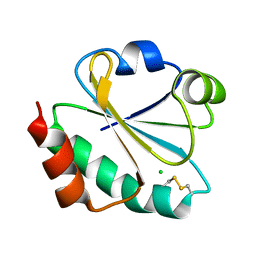 | | Crystal structure of ancestral thioredoxin, relative to the last common ancestor of the Cyanobacterial, Deinococcus and Thermus groups, LPBCA-L89K mutant. | | Descriptor: | CHLORIDE ION, LPBCA-L89K THIOREDOXIN | | Authors: | Gavira, J.A, Risso, V.A, Ibarra-Molero, B, Sanchez-Ruiz, J.M. | | Deposit date: | 2014-05-14 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mutational Studies on Resurrected Ancestral Proteins Reveal Conservation of Site-Specific Amino Acid Preferences Throughout Evolutionary History.
Mol.Biol.Evol., 32, 2015
|
|
4UHU
 
 | | W229D mutant of the last common ancestor of Gram-negative bacteria (GNCA) beta-lactamase class A | | Descriptor: | ACETATE ION, FORMIC ACID, GNCA LACTAMASE W229D | | Authors: | Gavira, J.A, Risso, V.A, Martinez-Rodriguez, S, Sanchez-Ruiz, J.M. | | Deposit date: | 2015-03-25 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.305 Å) | | Cite: | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
1ZZY
 
 | |
6YRS
 
 | | Structure of a new variant of GNCA ancestral beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gavira, J.A, Risso, V, Martinez-Rodriguez, S, Sanchez-Ruiz, J.M, Modi, T, Ozkan, S.B. | | Deposit date: | 2020-04-20 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hinge-shift mechanism as a protein design principle for the evolution of beta-lactamases from substrate promiscuity to specificity.
Nat Commun, 12, 2021
|
|
2K6R
 
 | | Protein folding on a highly rugged landscape: Experimental observation of glassy dynamics and structural frustration | | Descriptor: | Full Sequence Design 1 Synthetic Superstable | | Authors: | Sadqi, M, de Alba, E, Perez-Jimenez, R, Sanchez-Ruiz, J.M, Munoz, V. | | Deposit date: | 2008-07-18 | | Release date: | 2009-06-16 | | Last modified: | 2016-06-08 | | Method: | SOLUTION NMR | | Cite: | A designed protein as experimental model of primordial folding
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2YNX
 
 | | Crystal Structure of Ancestral Thioredoxin Relative to Last Archaea Common Ancestor (LACA) from the Precambrian Period | | Descriptor: | ACETATE ION, LACA THIOREDOXIN, SODIUM ION | | Authors: | Gavira, J.A, Ingles-Prieto, A, Ibarra-Molero, B, Sanchez-Ruiz, J.M. | | Deposit date: | 2012-10-19 | | Release date: | 2013-08-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Conservation of Protein Structure Over Four Billion Years
Structure, 21, 2013
|
|
2YOI
 
 | | Crystal Structure of Ancestral Thioredoxin Relative to Last Eukaryotes Common Ancestor (LECA) from the Precambrian Period | | Descriptor: | ACETATE ION, CHLORIDE ION, LECA THIOREDOXIN, ... | | Authors: | Gavira, J.A, Ingles-Prieto, A, Ibarra-Molero, B, Sanchez-Ruiz, J.M. | | Deposit date: | 2012-10-24 | | Release date: | 2013-08-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conservation of protein structure over four billion years.
Structure, 21, 2013
|
|
2YPM
 
 | |
2YN1
 
 | | Crystal Structure of Ancestral Thioredoxin Relative to Last Gamma- Proteobacteria Common Ancestor (LGPCA) from the Precambrian Period | | Descriptor: | LGPCA THIOREDOXIN, TRIETHYLENE GLYCOL | | Authors: | Gavira, J.A, Ingles-Prieto, A, Ibarra-Molero, B, Sanchez-Ruiz, J.M. | | Deposit date: | 2012-10-11 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conservation of Protein Structure Over Four Billion Years
Structure, 21, 2013
|
|
6TXD
 
 | | Variant W229D/F290W-12 of the last common ancestor of Gram-negative bacteria beta-lactamase class A (GNCA4) | | Descriptor: | ACETATE ION, Beta lactamase (GNCA4-12), FORMIC ACID, ... | | Authors: | Gavira, J.A, Risso, V, Sanchez-Ruiz, J.M, Romero-Rivera, A, Kamerlin, S.C.L. | | Deposit date: | 2020-01-14 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enhancing ade novoenzyme activity by computationally-focused ultra-low-throughput screening.
Chem Sci, 11, 2020
|
|
5FQK
 
 | | W229D and F290W mutant of the last common ancestor of Gram-negative bacteria (GNCA4) beta-lactamase class A bound to 5(6)-nitrobenzotriazole (TS-analog) | | Descriptor: | 6-NITROBENZOTRIAZOLE, GNCA4 LACTAMASE W229D AND F290W | | Authors: | Gavira, J.A, Risso, V.A, Martinez-Rodriguez, S, Sanchez-Ruiz, J.M. | | Deposit date: | 2015-12-11 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.767 Å) | | Cite: | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
5FQQ
 
 | | Last common ancestor of Gram-negative bacteria (GNCA4) beta-lactamase class A | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, DI(HYDROXYETHYL)ETHER, GNCA4 LACTAMASE | | Authors: | Gavira, J.A, Martinez-Rodriguez, S, Risso, V.A, Sanchez-Ruiz, J.M. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
6TY6
 
 | | Variant W229D/F290W-2 of the last common ancestor of Gram-negative bacteria beta-lactamase class A (GNCA4) bound to 5(6)-nitrobenzotriazole (TS-analog) | | Descriptor: | 6-NITROBENZOTRIAZOLE, ACETATE ION, Beta lactamase (GNCA4-2), ... | | Authors: | Gavira, J.A, Risso, V, Sanchez-Ruiz, J.M, Romero-Rivera, A, Kamerlin, S.C.L, Ortega-Munoz, M, Santoyo-Gonzalez, F. | | Deposit date: | 2020-01-15 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enhancing ade novoenzyme activity by computationally-focused ultra-low-throughput screening.
Chem Sci, 11, 2020
|
|
5FQI
 
 | | W229D and F290W mutant of the last common ancestor of Gram-negative bacteria (GNCA4) beta-lactamase class A | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gavira, J.A, Risso, V.A, Martinez-Rodriguez, S, Sanchez-Ruiz, J.M. | | Deposit date: | 2015-12-11 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
6TWW
 
 | | Variant W229D/F290W-19 of the last common ancestor of Gram-negative bacteria beta-lactamase class A (GNCA4) | | Descriptor: | ACETATE ION, Beta-Lactamase (GNCA4), FORMIC ACID, ... | | Authors: | Gavira, J.A, Risso, V, Sanchez-Ruiz, J.M, Romero-Rivera, A, Kamerlin, S.C.L. | | Deposit date: | 2020-01-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Enhancing ade novoenzyme activity by computationally-focused ultra-low-throughput screening.
Chem Sci, 11, 2020
|
|
5FQJ
 
 | | W229D mutant of the last common ancestor of Gram-negative bacteria (GNCA) beta-lactamase bound to 5(6)-nitrobenzotriazole (TS-analog) | | Descriptor: | 6-NITROBENZOTRIAZOLE, GNCA LACTAMASE W229D | | Authors: | Gavira, J.A, Martinez-Rodriguez, S, Risso, V.A, Sanchez-Ruiz, J.M. | | Deposit date: | 2015-12-11 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.271 Å) | | Cite: | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
4C75
 
 | | Consensus (ALL-CON) beta-lactamase class A | | Descriptor: | ACETATE ION, BETA-LACTAMASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gavira, J.A, Risso, V.A, Sanchez-Ruiz, J.M. | | Deposit date: | 2013-09-19 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Phenotypic Comparisons of Consensus Variants Versus Laboratory Resurrections of Precambrian Proteins.
Proteins, 82, 2014
|
|
4C6Y
 
 | | Ancestral PNCA (last common ancestors of Gram-positive and Gram- negative bacteria) beta-lactamase class A | | Descriptor: | ACETATE ION, BETA-LACTAMASE, CHLORIDE ION, ... | | Authors: | Gavira, J.A, Risso, V.A, Sanchez-Ruiz, J.M. | | Deposit date: | 2013-09-19 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Phenotypic Comparisons of Consensus Variants Versus Laboratory Resurrections of Precambrian Proteins.
Proteins, 82, 2014
|
|
2H74
 
 | |
2H75
 
 | |
2H73
 
 | |
2H6X
 
 | |
