8BPV
 
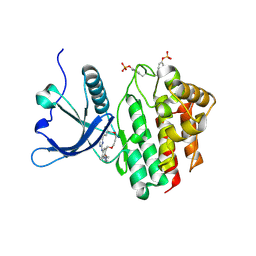 | | Crystal structure of JAK2 JH1 in complex with pacritinib | | Descriptor: | 11-(2-pyrrolidin-1-yl-ethoxy)-14,19-dioxa-5,7,26-triaza-tetracyclo[19.3.1.1(2,6).1(8,12)]heptacosa-1(25),2(26),3,5,8,10,12(27),16,21,23-decaene, Tyrosine-protein kinase JAK2 | | Authors: | Miao, Y, Haikarainen, T. | | Deposit date: | 2022-11-18 | | Release date: | 2023-11-29 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional and Structural Characterization of Clinical-Stage Janus Kinase 2 Inhibitors Identifies Determinants for Drug Selectivity.
J.Med.Chem., 67, 2024
|
|
8BM2
 
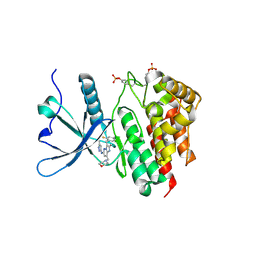 | | Crystal structure of JAK2 JH1 in complex with gandotinib | | Descriptor: | 3-[(4-chloranyl-2-fluoranyl-phenyl)methyl]-2-methyl-~{N}-(5-methyl-1~{H}-pyrazol-3-yl)-8-(morpholin-4-ylmethyl)imidazo[1,2-b]pyridazin-6-amine, Tyrosine-protein kinase JAK2 | | Authors: | Miao, Y, Haikarainen, T. | | Deposit date: | 2022-11-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Functional and Structural Characterization of Clinical-Stage Janus Kinase 2 Inhibitors Identifies Determinants for Drug Selectivity.
J.Med.Chem., 67, 2024
|
|
2B0K
 
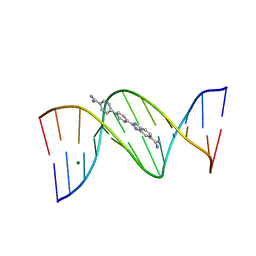 | | Crystal structure of the DB921-D(CGCGAATTCGCG)2 complex. | | Descriptor: | 2-(4'-AMIDINOBIPHENYL-4-YL)-1H-BENZIMIDAZOLE-5-AMIDINE, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | Authors: | Miao, Y, Lee, M.P, Parkinson, G.N, Batista-Parra, A, Ismail, M.A, Neidle, S, Boykin, D.W, Wilson, W.D. | | Deposit date: | 2005-09-14 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Out-of-Shape DNA Minor Groove Binders: Induced Fit Interactions of Heterocyclic Dications with the DNA Minor Groove.
Biochemistry, 44, 2005
|
|
8BPW
 
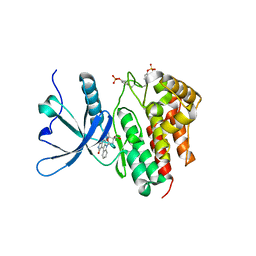 | | Crystal structure of JAK2 JH1 in complex with lestaurtinib | | Descriptor: | Lestaurtinib, Tyrosine-protein kinase JAK2 | | Authors: | Miao, Y, Haikarainen, T. | | Deposit date: | 2022-11-18 | | Release date: | 2023-11-29 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional and Structural Characterization of Clinical-Stage Janus Kinase 2 Inhibitors Identifies Determinants for Drug Selectivity.
J.Med.Chem., 67, 2024
|
|
5HUS
 
 | |
5HUU
 
 | | Structure of Candida albicans trehalose-6-phosphate synthase in complex with UDP and glucose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, Alpha,alpha-trehalose-phosphate synthase [UDP-forming], URIDINE-5'-DIPHOSPHATE | | Authors: | Miao, Y, Brennan, R.G. | | Deposit date: | 2016-01-27 | | Release date: | 2017-05-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural and In Vivo Studies on Trehalose-6-Phosphate Synthase from Pathogenic Fungi Provide Insights into Its Catalytic Mechanism, Biological Necessity, and Potential for Novel Antifungal Drug Design.
MBio, 8, 2017
|
|
5HVM
 
 | | Structure of Aspergillus fumigatus trehalose-6-phosphate synthase A in complex with UDP and validoxylamine A | | Descriptor: | (1S,2S,3R,6S)-4-(HYDROXYMETHYL)-6-{[(1S,2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-(HYDROXYMETHYL)CYCLOHEXYL]AMINO}CYCLOHEX-4-ENE-1,2,3-TRIOL, Alpha,alpha-trehalose-phosphate synthase (UDP-forming), URIDINE-5'-DIPHOSPHATE | | Authors: | Miao, Y, Brennan, R.G. | | Deposit date: | 2016-01-28 | | Release date: | 2017-05-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.815 Å) | | Cite: | Structural and In Vivo Studies on Trehalose-6-Phosphate Synthase from Pathogenic Fungi Provide Insights into Its Catalytic Mechanism, Biological Necessity, and Potential for Novel Antifungal Drug Design.
MBio, 8, 2017
|
|
5HUT
 
 | | Structure of Candida albicans trehalose-6-phosphate synthase in complex with UDP-glucose | | Descriptor: | Alpha,alpha-trehalose-phosphate synthase [UDP-forming], PENTAETHYLENE GLYCOL, SODIUM ION, ... | | Authors: | Miao, Y, Brennan, R.G. | | Deposit date: | 2016-01-27 | | Release date: | 2017-05-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and In Vivo Studies on Trehalose-6-Phosphate Synthase from Pathogenic Fungi Provide Insights into Its Catalytic Mechanism, Biological Necessity, and Potential for Novel Antifungal Drug Design.
MBio, 8, 2017
|
|
5HVO
 
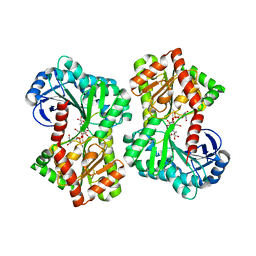 | | Structure of Aspergillus fumigatus trehalose-6-phosphate synthase B in complex with UDP and validoxylamine A | | Descriptor: | (1S,2S,3R,6S)-4-(HYDROXYMETHYL)-6-{[(1S,2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-(HYDROXYMETHYL)CYCLOHEXYL]AMINO}CYCLOHEX-4-ENE-1,2,3-TRIOL, Alpha,alpha-trehalose-phosphate synthase (UDP-forming), URIDINE-5'-DIPHOSPHATE | | Authors: | Miao, Y, Brennan, R.G. | | Deposit date: | 2016-01-28 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.467 Å) | | Cite: | Structural and In Vivo Studies on Trehalose-6-Phosphate Synthase from Pathogenic Fungi Provide Insights into Its Catalytic Mechanism, Biological Necessity, and Potential for Novel Antifungal Drug Design.
MBio, 8, 2017
|
|
5HVL
 
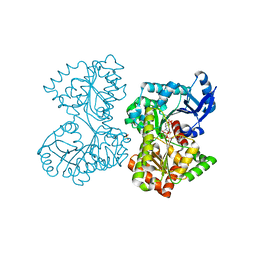 | | Structure of Candida albicans trehalose-6-phosphate synthase in complex with UDP and validoxylamine A | | Descriptor: | (1S,2S,3R,6S)-4-(HYDROXYMETHYL)-6-{[(1S,2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-(HYDROXYMETHYL)CYCLOHEXYL]AMINO}CYCLOHEX-4-ENE-1,2,3-TRIOL, Alpha,alpha-trehalose-phosphate synthase [UDP-forming], SULFATE ION, ... | | Authors: | Miao, Y, Brennan, R.G. | | Deposit date: | 2016-01-28 | | Release date: | 2017-05-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Structural and In Vivo Studies on Trehalose-6-Phosphate Synthase from Pathogenic Fungi Provide Insights into Its Catalytic Mechanism, Biological Necessity, and Potential for Novel Antifungal Drug Design.
MBio, 8, 2017
|
|
5HUV
 
 | |
5DXN
 
 | |
5DXI
 
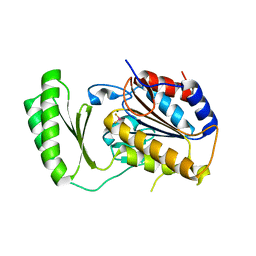 | | Structure of C. albicans Trehalose-6-phosphate phosphatase C-terminal domain | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, ... | | Authors: | Miao, Y, Brennan, R.G. | | Deposit date: | 2015-09-23 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of trehalose-6-phosphate phosphatase from pathogenic fungi reveal the mechanisms of substrate recognition and catalysis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
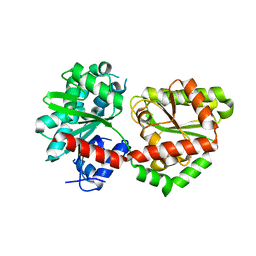
5DXF
 
 | |
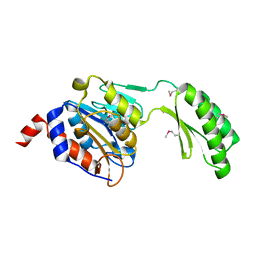
5DXL
 
 | |
5DX9
 
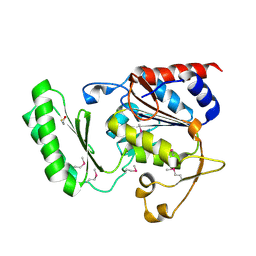 | | Structure of trehalose-6-phosphate phosphatase from Cryptococcus neoformans | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, BETA-MERCAPTOETHANOL, MAGNESIUM ION, ... | | Authors: | Miao, Y, Brennan, R.G. | | Deposit date: | 2015-09-23 | | Release date: | 2016-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of trehalose-6-phosphate phosphatase from pathogenic fungi reveal the mechanisms of substrate recognition and catalysis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5DXO
 
 | |
7R84
 
 | |
7R85
 
 | | Structure of mouse Bai1 (ADGRB1) TSR3 domain | | Descriptor: | Vasculostatin-120, alpha-D-mannopyranose, beta-D-glucopyranose-(1-3)-alpha-L-fucopyranose | | Authors: | Miao, Y, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-06-26 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | RTN4/NoGo-receptor binding to BAI adhesion-GPCRs regulates neuronal development.
Cell, 184, 2021
|
|
7R86
 
 | | Structure of mouse BAI1 (ADGRB1) in complex with mouse Nogo receptor (RTN4R) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-AMINOBENZOIC ACID, ... | | Authors: | Miao, Y, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-06-26 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | RTN4/NoGo-receptor binding to BAI adhesion-GPCRs regulates neuronal development.
Cell, 184, 2021
|
|
6NE1
 
 | | Designed repeat protein in complex with Fz4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Frizzled-4, Pfs, ... | | Authors: | Miao, Y, Jude, K.M, Garcia, K.C. | | Deposit date: | 2018-12-15 | | Release date: | 2019-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Receptor subtype discrimination using extensive shape complementary designed interfaces.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6NE4
 
 | | Designed repeat protein specifically in complex with Fz7CRD | | Descriptor: | 1,2-ETHANEDIOL, Designed repeat binding protein, Frizzled-7, ... | | Authors: | Miao, Y, Jude, K.M, Garcia, K.C. | | Deposit date: | 2018-12-16 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Receptor subtype discrimination using extensive shape complementary designed interfaces.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6NDZ
 
 | | Designed repeat protein in complex with Fz8 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ACETIC ACID, ... | | Authors: | Miao, Y, Jude, K.M, Garcia, K.C. | | Deposit date: | 2018-12-14 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.264 Å) | | Cite: | Receptor subtype discrimination using extensive shape complementary designed interfaces.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6NE2
 
 | | Designed repeat protein in complex with Fz7 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Miao, Y, Jude, K.M, Garcia, K.C. | | Deposit date: | 2018-12-15 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Receptor subtype discrimination using extensive shape complementary designed interfaces.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3N6M
 
 | | Crystal structure of EV71 RdRp in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, NICKEL (II) ION, RNA-dependent RNA polymerase | | Authors: | Wu, Y, Lou, Z.Y, Miao, Y, Yu, Y, Rao, Z.H. | | Deposit date: | 2010-05-26 | | Release date: | 2011-06-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of EV71 RNA-dependent RNA polymerase in complex with substrate and analogue provide a drug target against the hand-foot-and-mouth disease pandemic in China.
Protein Cell, 1, 2010
|
|
