3MPH
 
 | | The structure of human diamine oxidase complexed with an inhibitor aminoguanidine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Amiloride-sensitive amine oxidase, CALCIUM ION, ... | | Authors: | McGrath, A.P, Guss, J.M. | | Deposit date: | 2010-04-27 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Correlation of active site metal content in human diamine oxidase with trihydroxyphenylalanine quinone cofactor biogenesis
Biochemistry, 49, 2010
|
|
3PGB
 
 | | Crystal structure of Aspergillus nidulans amine oxidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | McGrath, A.P, Guss, J.M. | | Deposit date: | 2010-11-01 | | Release date: | 2011-07-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure and Activity of Aspergillus nidulans Copper Amine Oxidase
Biochemistry, 50, 2011
|
|
3HI7
 
 | | Crystal structure of human diamine oxidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Amiloride-sensitive amine oxidase, CALCIUM ION, ... | | Authors: | McGrath, A.P, Guss, J.M. | | Deposit date: | 2009-05-19 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Structure and inhibition of human diamine oxidase
Biochemistry, 48, 2009
|
|
3K5T
 
 | | Crystal structure of human diamine oxidase in space group C2221 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | McGrath, A.P, Guss, J.M. | | Deposit date: | 2009-10-08 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | A new crystal form of human diamine oxidase.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3HIG
 
 | |
3HII
 
 | |
4PW9
 
 | |
4PW3
 
 | |
7SQO
 
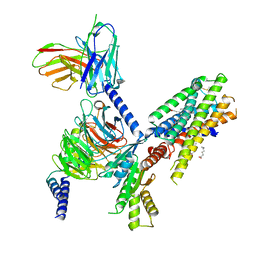 | | Structure of the orexin-2 receptor(OX2R) bound to TAK-925, Gi and scFv16 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | McGrath, A.P, Kang, Y, Flinspach, M. | | Deposit date: | 2021-11-05 | | Release date: | 2022-05-25 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Molecular mechanism of the wake-promoting agent TAK-925.
Nat Commun, 13, 2022
|
|
4XWK
 
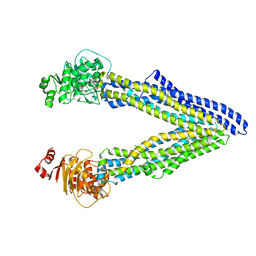 | | P-glycoprotein co-crystallized with BDE-100 | | Descriptor: | 2,4-dibromophenyl 2,4,6-tribromophenyl ether, Multidrug resistance protein 1A | | Authors: | McGrath, A.P, Rees, S.D, Chang, G. | | Deposit date: | 2015-01-28 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Global marine pollutants inhibit P-glycoprotein: Environmental levels, inhibitory effects, and cocrystal structure.
Sci Adv, 2, 2016
|
|
4Q9J
 
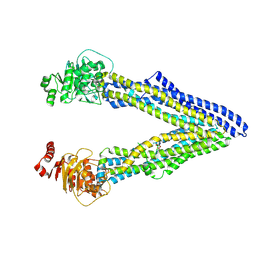 | | P-glycoprotein cocrystallised with QZ-Val | | Descriptor: | Multidrug resistance protein 1A, N-{[5-AMINO-1-(5-O-PHOSPHONO-BETA-D-ARABINOFURANOSYL)-1H-IMIDAZOL-4-YL]CARBONYL}-L-ASPARTIC ACID | | Authors: | McGrath, A.P, Szewczyk, P, Chang, G. | | Deposit date: | 2014-05-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4Q9H
 
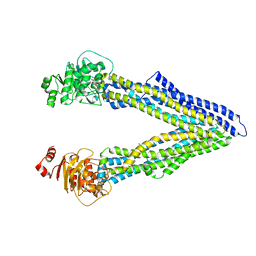 | | P-glycoprotein at 3.4 A resolution | | Descriptor: | Multidrug resistance protein 1A | | Authors: | McGrath, A.P, Szewczyk, P, Chang, G. | | Deposit date: | 2014-05-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4PWA
 
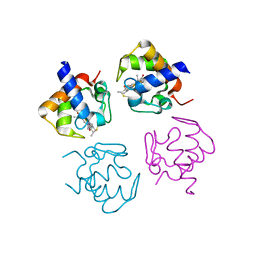 | |
4Q9K
 
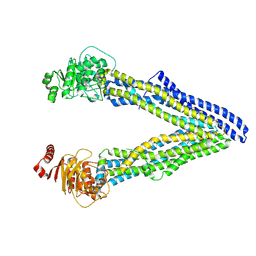 | | P-glycoprotein cocrystallised with QZ-Leu | | Descriptor: | (30F)L(30F)L(30F)L Peptide, Multidrug resistance protein 1A | | Authors: | McGrath, A.P, Szewczyk, P, Chang, G. | | Deposit date: | 2014-05-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4Q9I
 
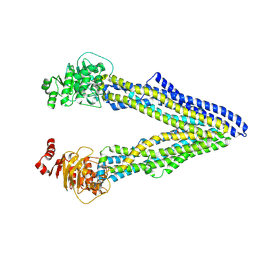 | | P-glycoprotein cocrystallised with QZ-Ala | | Descriptor: | (30F)A(30F)A(30F)A Peptide, Multidrug resistance protein 1A | | Authors: | McGrath, A.P, Szewczyk, P, Chang, G. | | Deposit date: | 2014-05-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.781 Å) | | Cite: | Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4Q9L
 
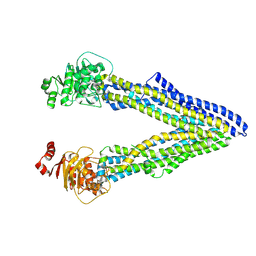 | | P-glycoprotein cocrystallised with QZ-Phe | | Descriptor: | (30F)F(30F)F(30F)F Peptide, Multidrug resistance protein 1A | | Authors: | McGrath, A.P, Szewczyk, P, Chang, G. | | Deposit date: | 2014-05-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3W5I
 
 | |
3W5J
 
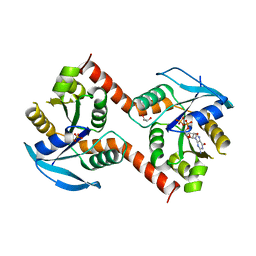 | | Crystal structure of GDP-bound NfeoB from Gallionella capsiferriformans | | Descriptor: | Ferrous iron transport protein B, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Deshpande, C.N, McGrath, A.P, Jormakka, M. | | Deposit date: | 2013-01-30 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Structure of an atypical FeoB G-domain reveals a putative domain-swapped dimer.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3ALA
 
 | | Crystal structure of vascular adhesion protein-1 in space group C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ernberg, K.E, McGrath, A.P, Guss, J.M. | | Deposit date: | 2010-07-29 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A new crystal form of human vascular adhesion protein 1
Acta Crystallogr.,Sect.F, 66, 2010
|
|
6OCE
 
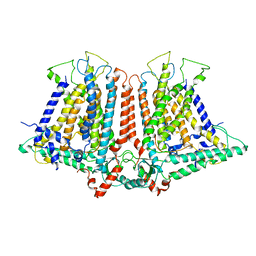 | | Structure of the rice hyperosmolality-gated ion channel OSCA1.2 | | Descriptor: | stress-gated cation channel 1.2 | | Authors: | Maity, K, Heumann, J.M, McGrath, A.P, Chang, G, Stowell, M.H. | | Deposit date: | 2019-03-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cryo-EM structure of OSCA1.2 fromOryza sativaelucidates the mechanical basis of potential membrane hyperosmolality gating.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3TU6
 
 | | The Structure of a Pseudoazurin From Sinorhizobium meliltoi | | Descriptor: | COPPER (II) ION, GLYCEROL, Pseudoazurin (Blue copper protein) | | Authors: | Laming, E.M, McGrath, A.P, Guss, J.M, Maher, M.J. | | Deposit date: | 2011-09-16 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray crystal structure of a pseudoazurin from Sinorhizobium meliloti.
J.Inorg.Biochem., 115, 2012
|
|
3G9Y
 
 | | Crystal structure of the second zinc finger from ZRANB2/ZNF265 bound to 6 nt ssRNA sequence AGGUAA | | Descriptor: | RNA (5'-R(*AP*GP*GP*UP*AP*A)-3'), ZINC ION, Zinc finger Ran-binding domain-containing protein 2 | | Authors: | Loughlin, F.E, McGrath, A.P, Lee, M, Guss, J.M, Mackay, J.P. | | Deposit date: | 2009-02-15 | | Release date: | 2009-03-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The zinc fingers of the SR-like protein ZRANB2 are single-stranded RNA-binding domains that recognize 5' splice site-like sequences
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7JY4
 
 | | hALK in complex with ((1S,2S)-1-(2,4-difluorophenyl)-2-(2-(3-methyl-1H-pyrazol-5-yl)-4-(trifluoromethyl)phenoxy)cyclopropyl)methanamine | | Descriptor: | 1-{(1S,2S)-1-(2,4-difluorophenyl)-2-[2-(3-methyl-1H-pyrazol-5-yl)-4-(trifluoromethyl)phenoxy]cyclopropyl}methanamine, ALK tyrosine kinase receptor | | Authors: | McGrath, A.P, Zou, H, Lane, W, Saikatendu, K. | | Deposit date: | 2020-08-28 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Discovery of Novel and Highly Selective Cyclopropane ALK Inhibitors through a Fragment-Assisted, Structure-Based Drug Design.
Acs Omega, 5, 2020
|
|
7JYR
 
 | | hALK in complex with 1-[(1R,2R)-1-(2,4-difluorophenyl)-2-[2-(5-methyl-1H-pyrazol-3-yl)-4-(trifluoromethyl)phenoxy]cyclopropyl]methanamine | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(1R,2R)-2-(2,4-difluorophenyl)cyclopropyl]oxy}-3-(5-methyl-1H-pyrazol-3-yl)benzonitrile, ALK tyrosine kinase receptor | | Authors: | McGrath, A.P, Zou, H, Lane, W, Saikatendu, K. | | Deposit date: | 2020-08-31 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Discovery of Novel and Highly Selective Cyclopropane ALK Inhibitors through a Fragment-Assisted, Structure-Based Drug Design.
Acs Omega, 5, 2020
|
|
7JYS
 
 | | hALK in complex with 3-(3-chlorophenyl)-5-methyl-1H-pyrazole | | Descriptor: | 1,2-ETHANEDIOL, 3-(3-chlorophenyl)-5-methyl-1H-pyrazole, ALK tyrosine kinase receptor | | Authors: | McGrath, A.P, Zou, H, Lane, W, Saikatendu, K. | | Deposit date: | 2020-08-31 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery of Novel and Highly Selective Cyclopropane ALK Inhibitors through a Fragment-Assisted, Structure-Based Drug Design.
Acs Omega, 5, 2020
|
|
