4NB5
 
 | |
5D9R
 
 | | Crystal structure of a conserved domain in the intermembrane space region of the plastid division protein ARC6 | | Descriptor: | Protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6, chloroplastic | | Authors: | Radhakrishnan, A, Kumar, N, Su, C.-C, Chou, T.-H, Yu, E. | | Deposit date: | 2015-08-18 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Crystal structure of a conserved domain in the intermembrane space region of the plastid division protein ARC6.
Protein Sci., 25, 2016
|
|
5V4F
 
 | | Crystal Structure of the Protein of Unknown Function of the Conserved Rid Protein Family YyfB from Yersinia pestis | | Descriptor: | GLYCEROL, Putative translational inhibitor protein | | Authors: | Kim, Y, Chhor, G, Endres, M, Krishnan, A, Babnigg, G, Schneewind, O, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-03-09 | | Release date: | 2017-04-05 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Crystal Structure of the Protein of Unknown Function of the Conserved Rid Protein Family YyfB from Yersinia pestis
To Be Published
|
|
5V4D
 
 | | Crystal Structure of the Protein of Unknown Function of the Conserved Rid Protein Family YyfA from Yersinia pestis | | Descriptor: | ACETIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Krishnan, A, Babnigg, G, Schneewind, O, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-03-09 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Protein of Unknown Function of the Conserved Rid Protein Family YyfA from Yersinia pestis
To Be Published
|
|
6MYK
 
 | | Pleurotus ostreatus OstreolysinA mutant E69A with Bis-Tris | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Ostreolysin A6, ... | | Authors: | Tomchick, D.R, Radhakrishnan, A, Endapally, S. | | Deposit date: | 2018-11-01 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Discrimination between Two Conformations of Sphingomyelin in Plasma Membranes.
Cell, 176, 2019
|
|
6MYJ
 
 | | Pleurotus ostreatus OstreolysinA plus sphingomyelin | | Descriptor: | 1,2-ETHANEDIOL, N-[(2S)-1-hydroxypropan-2-yl]butanamide, Ostreolysin A6, ... | | Authors: | Tomchick, D.R, Radhakrishnan, A, Endapally, S. | | Deposit date: | 2018-11-01 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Molecular Discrimination between Two Conformations of Sphingomyelin in Plasma Membranes.
Cell, 176, 2019
|
|
6MYI
 
 | | Pleurotus ostreatus OstreolysinA | | Descriptor: | 1,2-ETHANEDIOL, Ostreolysin A6, SODIUM ION | | Authors: | Tomchick, D.R, Radhakrishnan, A, Endapally, S. | | Deposit date: | 2018-11-01 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Molecular Discrimination between Two Conformations of Sphingomyelin in Plasma Membranes.
Cell, 176, 2019
|
|
6BHP
 
 | |
7LKF
 
 | | WT Chicken Scap L1-L7 / Fab 4G10 complex focused refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4G10 heavy chain, 4G10 light chain, ... | | Authors: | Kober, D.L, Radhakrishnan, A, Goldstein, J.L, Brown, M.S, Clark, L.D, Bai, X.-C, Rosenbaum, D.M. | | Deposit date: | 2021-02-02 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Scap structures highlight key role for rotation of intertwined luminal loops in cholesterol sensing.
Cell, 184, 2021
|
|
7LKH
 
 | | Chicken Scap D435V L1-L7 domain / Fab complex focused map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4G10 Fab heavy chain, 4G10 Fab kappa chain, ... | | Authors: | Kober, D.L, Radhakrishnan, A, Goldstein, J.L, Brown, M.S, Clark, L.D, Bai, X.-C, Rosenbaum, D.M. | | Deposit date: | 2021-02-02 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Scap structures highlight key role for rotation of intertwined luminal loops in cholesterol sensing.
Cell, 184, 2021
|
|
2LKM
 
 | |
7XKA
 
 | | Structure of human beta2 adrenergic receptor bound to constrained epinephrine | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Camelid Antibody Fragment, Endolysin,Beta-2 adrenergic receptor, ... | | Authors: | Xu, X, Shonberg, J, Kaindl, J, Clark, M, Stobel, A, Maul, L, Mayer, D, Hubner, H, Venkatakrishnan, A, Dror, R, Kobilka, B.K, Sunahara, R, Liu, X, Gmeiner, P. | | Deposit date: | 2022-04-19 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Constrained catecholamines gain beta 2 AR selectivity through allosteric effects on pocket dynamics.
Nat Commun, 14, 2023
|
|
7XK9
 
 | | Structure of human beta2 adrenergic receptor bound to constrained isoproterenol | | Descriptor: | (5R,6R)-6-(propan-2-ylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Camelid Antibody Fragment, Endolysin,Beta-2 adrenergic receptor, ... | | Authors: | Xu, X, Shonberg, J, Kaindl, J, Clark, M, Stobel, A, Maul, L, Mayer, D, Hubner, H, Venkatakrishnan, A, Dror, R, Kobilka, B.K, Sunahara, R, Liu, X, Gmeiner, P. | | Deposit date: | 2022-04-19 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Constrained catecholamines gain beta 2 AR selectivity through allosteric effects on pocket dynamics.
Nat Commun, 14, 2023
|
|
8BEO
 
 | | Crystal structure of E. coli glyoxylate carboligase mutant I393A with MAP | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2,3-DIMETHOXY-5-METHYL-1,4-BENZOQUINONE, ... | | Authors: | Shaanan, B, Binshtein, E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of E. coli glyoxylate carboligase mutant I393A with MAP
To Be Published
|
|
6QSM
 
 | |
6QSO
 
 | |
6QSL
 
 | |
4R1I
 
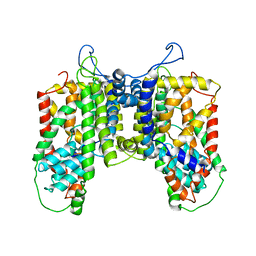 | |
3F6E
 
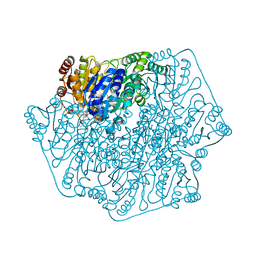 | | Crystal structure of benzoylformate decarboxylase in complex with the pyridyl inhibitor 3-PKB | | Descriptor: | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-2-[(1S,2E)-1-hydroxy-3-pyridin-3-ylprop-2-en-1-yl]-4-methyl-1,3-thiazol-3-ium, Benzoylformate decarboxylase, MAGNESIUM ION | | Authors: | Brandt, G.S, McLeish, M.J, Kenyon, G.L, Petsko, G.A, Ringe, D, Jordan, F. | | Deposit date: | 2008-11-05 | | Release date: | 2008-12-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Detection and time course of formation of major thiamin diphosphate-bound covalent intermediates derived from a chromophoric substrate analogue on benzoylformate decarboxylase.
Biochemistry, 48, 2009
|
|
3F6B
 
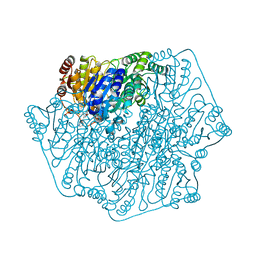 | | Crystal structure of benzoylformate decarboxylase in complex with the pyridyl inhibitor PAA | | Descriptor: | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-2-[(1S,2E)-1-hydroxy-3-pyridin-3-ylprop-2-en-1-yl]-4-methyl-1,3-thiazol-3-ium, Benzoylformate decarboxylase, MAGNESIUM ION | | Authors: | Brandt, G.S, McLeish, M.J, Kenyon, G.L, Petsko, G.A, Ringe, D, Jordan, F. | | Deposit date: | 2008-11-05 | | Release date: | 2008-12-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Detection and time course of formation of major thiamin diphosphate-bound covalent intermediates derived from a chromophoric substrate analogue on benzoylformate decarboxylase.
Biochemistry, 48, 2009
|
|
4XU6
 
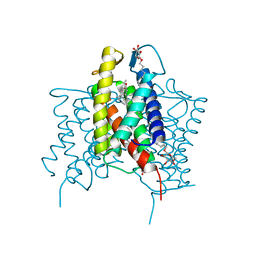 | | Crystal structure of cross-linked MvINS R77C trimer at 1.9A resolution | | Descriptor: | N-TRIDECANOIC ACID, Uncharacterized protein, octyl beta-D-glucopyranoside | | Authors: | Ren, R.B, Wu, J.P, Yan, C.Y, He, Y, Yan, N. | | Deposit date: | 2015-01-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | PROTEIN STRUCTURE. Crystal structure of a mycobacterial Insig homolog provides insight into how these sensors monitor sterol levels
Science, 349, 2015
|
|
8VDJ
 
 | | Crystal structure of SARS-CoV-2 3CL protease (3CLpro) as a covalent complex with EDP-235 | | Descriptor: | 3C-like proteinase nsp5, 4,6,7-trifluoro-N-{(2S)-1-[(3R,5'R)-5'-(iminomethyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidin]-1'-yl]-4-methyl-1-oxopentan-2-yl}-N-methyl-1H-indole-2-carboxamide, THIOCYANATE ION | | Authors: | Cade, I.A, Rhodin, M.H.J. | | Deposit date: | 2023-12-15 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | The small molecule inhibitor of SARS-CoV-2 3CLpro EDP-235 prevents viral replication and transmission in vivo.
Nat Commun, 15, 2024
|
|
2M2D
 
 | | Human programmed cell death 1 receptor | | Descriptor: | Programmed cell death protein 1 | | Authors: | Veverka, V, Cheng, X, Waters, L.C, Muskett, F.W, Morgan, S, Lesley, A, Griffiths, M, Stubberfield, C, Griffin, R, Henry, A.J, Robinson, M.K, Jansson, A, Ladbury, J.E, Ikemizu, S, Davis, S.J, Carr, M.D. | | Deposit date: | 2012-12-18 | | Release date: | 2013-02-27 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure and interactions of the human programmed cell death 1 receptor.
J.Biol.Chem., 288, 2013
|
|
5KHS
 
 | |
5KHN
 
 | |
