6KRE
 
 | | TRiC at 0.05 mM ADP-AlFx, Conformation 2, 0.05-C2 | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-21 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.45 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KRD
 
 | | TRiC at 0.05 mM ADP-AlFx, Conformation 4, 0.05-C4 | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-21 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.38 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KS8
 
 | | TRiC at 0.1 mM ADP-AlFx, Conformation 4, 0.1-C4 | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-23 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KS7
 
 | | TRiC at 0.1 mM ADP-AlFx, Conformation 1, 0.1-C1 | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-23 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.62 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KS6
 
 | | TRiC at 0.2 mM ADP-AlFx, Conformation 1, 0.2-C1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-23 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4ID7
 
 | | ACK1 kinase in complex with the inhibitor cis-3-[8-amino-1-(4-phenoxyphenyl)imidazo[1,5-a]pyrazin-3-yl]cyclobutanol | | Descriptor: | Activated CDC42 kinase 1, SULFATE ION, cis-3-[8-amino-1-(4-phenoxyphenyl)imidazo[1,5-a]pyrazin-3-yl]cyclobutanol | | Authors: | Jin, M, Wang, J, Kleinberg, A, Kadalbajoo, M, Siu, K, Cooke, A, Bittner, M, Yao, Y, Thelemann, A, Ji, Q, Bhagwat, S, Crew, A.P, Pachter, J, Epstein, D, Mulvihill, M.J. | | Deposit date: | 2012-12-11 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of potent, selective and orally bioavailable imidazo[1,5-a]pyrazine derived ACK1 inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
8VS6
 
 | | L-TGF-b3/avb8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Jin, M, Cheng, Y, Nishimura, S.L. | | Deposit date: | 2024-01-23 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Dynamic allostery drives autocrine and paracrine TGF-beta signaling.
Cell, 2024
|
|
8VSD
 
 | | avb8/L-TGF-b1/GARP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Jin, M, Cheng, Y, Nishimura, S.L. | | Deposit date: | 2024-01-23 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Dynamic allostery drives autocrine and paracrine TGF-beta signaling.
Cell, 2024
|
|
8VSC
 
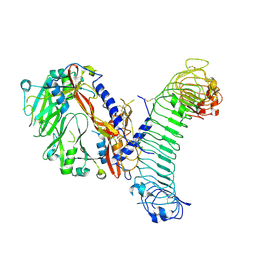 | | L-TGF-b1/GARP | | Descriptor: | Transforming growth factor beta activator LRRC32, Transforming growth factor beta-1 proprotein | | Authors: | Jin, M, Cheng, Y, Nishimura, S.L. | | Deposit date: | 2024-01-23 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Dynamic allostery drives autocrine and paracrine TGF-beta signaling.
Cell, 2024
|
|
8VSB
 
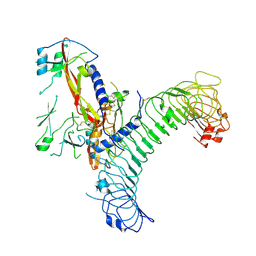 | | L-TGF-b3/GARP | | Descriptor: | Transforming growth factor beta activator LRRC32, Transforming growth factor beta-3 proprotein | | Authors: | Jin, M, Cheng, Y, Nishimura, S.L. | | Deposit date: | 2024-01-23 | | Release date: | 2024-09-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Dynamic allostery drives autocrine and paracrine TGF-beta signaling.
Cell, 2024
|
|
4CMQ
 
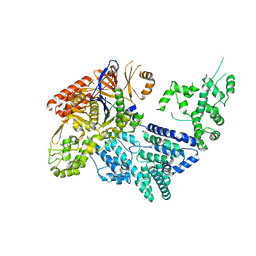 | | Crystal structure of Mn-bound S.pyogenes Cas9 | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MANGANESE (II) ION, SULFATE ION | | Authors: | Jinek, M, Jiang, F, Taylor, D.W, Sternberg, S.H, Kaya, E, Ma, E, Anders, C, Hauer, M, Zhou, K, Lin, S, Kaplan, M, Iavarone, A.T, Charpentier, E, Nogales, E, Doudna, J.A. | | Deposit date: | 2014-01-17 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structures of Cas9 Endonucleases Reveal RNA- Mediated Conformational Activation
Science, 343, 2014
|
|
1TON
 
 | |
4CMP
 
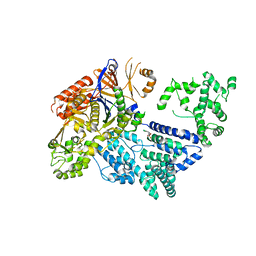 | | Crystal structure of S. pyogenes Cas9 | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, SULFATE ION | | Authors: | Jinek, M, Jiang, F, Taylor, D.W, Sternberg, S.H, Kaya, E, Ma, E, Anders, C, Hauer, M, Zhou, K, Lin, S, Kaplan, M, Iavarone, A.T, Charpentier, E, Nogales, E, Doudna, J.A. | | Deposit date: | 2014-01-16 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation.
Science, 343, 2014
|
|
1CHO
 
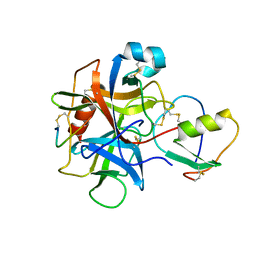 | | CRYSTAL AND MOLECULAR STRUCTURES OF THE COMPLEX OF ALPHA-*CHYMOTRYPSIN WITH ITS INHIBITOR TURKEY OVOMUCOID THIRD DOMAIN AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | ALPHA-CHYMOTRYPSIN A, TURKEY OVOMUCOID THIRD DOMAIN (OMTKY3) | | Authors: | Fujinaga, M, Sielecki, A.R, Read, R.J, Ardelt, W, Laskowskijunior, M, James, M.N.G. | | Deposit date: | 1988-03-04 | | Release date: | 1988-07-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal and molecular structures of the complex of alpha-chymotrypsin with its inhibitor turkey ovomucoid third domain at 1.8 A resolution.
J.Mol.Biol., 195, 1987
|
|
1QRP
 
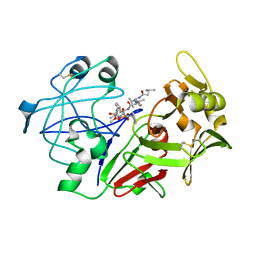 | | Human pepsin 3A in complex with a phosphonate inhibitor IVA-VAL-VAL-LEU(P)-(O)PHE-ALA-ALA-OME | | Descriptor: | PEPSIN 3A, methyl N-[(2S)-2-({(S)-hydroxy[(1R)-3-methyl-1-{[N-(3-methylbutanoyl)-L-valyl-L-valyl]amino}butyl]phosphoryl}oxy)-3-phenylpropanoyl]-L-alanyl-L-alaninate | | Authors: | Fujinaga, M, Cherney, M.M, Tarasova, N.I, Bartlett, P.A, Hanson, J.E, James, M.N.G. | | Deposit date: | 1999-06-15 | | Release date: | 1999-06-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural study of the complex between human pepsin and a phosphorus-containing peptidic -transition-state analog.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1W3B
 
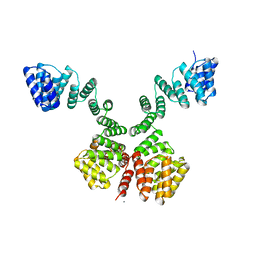 | | The superhelical TPR domain of O-linked GlcNAc transferase reveals structural similarities to importin alpha. | | Descriptor: | CALCIUM ION, UDP-N-ACETYLGLUCOSAMINE--PEPTIDE N-ACETYLGLUCOSAMINYLTRANSFERASE 110 | | Authors: | Jinek, M, Rehwinkel, J, Lazarus, B.D, Izaurralde, E, Hanover, J.A, Conti, E. | | Deposit date: | 2004-07-14 | | Release date: | 2004-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Superhelical Tpr-Repeat Domain of O-Linked Glcnac Transferase Exhibits Structural Similarities to Importin Alpha
Nat.Struct.Mol.Biol., 11, 2004
|
|
1SRY
 
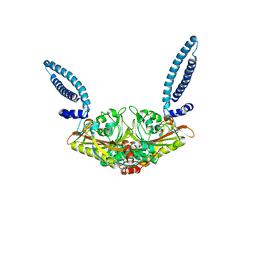 | |
1FUJ
 
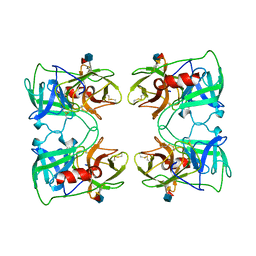 | | PR3 (MYELOBLASTIN) | | Descriptor: | PR3, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Fujinaga, M, Chernaia, M.M, Halenbeck, R, Koths, K, James, M.N.G. | | Deposit date: | 1996-01-25 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of PR3, a neutrophil serine proteinase antigen of Wegener's granulomatosis antibodies.
J.Mol.Biol., 261, 1996
|
|
1PSN
 
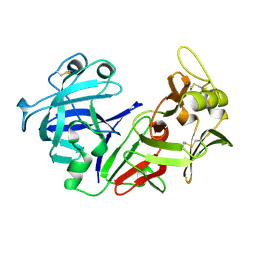 | | THE CRYSTAL STRUCTURE OF HUMAN PEPSIN AND ITS COMPLEX WITH PEPSTATIN | | Descriptor: | PEPSIN 3A | | Authors: | Fujinaga, M, Chernaia, M.M, Tarasova, N, Mosimann, S.C, James, M.N.G. | | Deposit date: | 1995-01-23 | | Release date: | 1995-04-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human pepsin and its complex with pepstatin.
Protein Sci., 4, 1995
|
|
1PSO
 
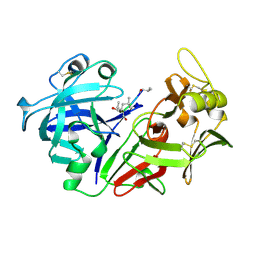 | | The crystal structure of human pepsin and its complex with pepstatin | | Descriptor: | PEPSIN 3A, PEPSTATIN | | Authors: | Fujinaga, M, Chernaia, M.M, Tarasova, N, Mosimann, S.C, James, M.N.G. | | Deposit date: | 1995-01-23 | | Release date: | 1995-04-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human pepsin and its complex with pepstatin.
Protein Sci., 4, 1995
|
|
1S2B
 
 | | Structure of SCP-B the first member of the Eqolisin family of Peptidases to have its structure determined | | Descriptor: | Scytalidopepsin B | | Authors: | Fujinaga, M, Cherney, M.M, Oyama, H, Oda, K, James, M.N. | | Deposit date: | 2004-01-08 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The molecular structure and catalytic mechanism of a novel carboxyl peptidase from Scytalidium lignicolum
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2Y35
 
 | | Crystal structure of Xrn1-substrate complex | | Descriptor: | DT11 (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP)-3', LD22664P, MAGNESIUM ION | | Authors: | Jinek, M, Coyle, S.M, Doudna, J.A. | | Deposit date: | 2010-12-18 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Coupled 5' Nucleotide Recognition and Processivity in Xrn1-Mediated Mrna Decay.
Mol.Cell, 41, 2011
|
|
2X04
 
 | | Crystal structure of the PABC-TNRC6C complex | | Descriptor: | POLYADENYLATE-BINDING PROTEIN 1, SULFATE ION, TRINUCLEOTIDE REPEAT-CONTAINING GENE 6C PROTEIN | | Authors: | Jinek, M, Fabian, M.R, Coyle, S.M, Sonenberg, N, Doudna, J.A. | | Deposit date: | 2009-12-04 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Insights Into the Human Gw182-Pabc Interaction in Microrna-Mediated Deadenylation
Nat.Struct.Mol.Biol., 17, 2010
|
|
2VXG
 
 | | Crystal structure of the conserved C-terminal region of Ge-1 | | Descriptor: | CG6181-PA, ISOFORM A | | Authors: | Jinek, M, Eulalio, A, Lingel, A, Helms, S, Conti, E, Izaurralde, E. | | Deposit date: | 2008-07-04 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The C-Terminal Region of Ge-1 Presents Conserved Structural Features Required for P-Body Localization.
RNA, 14, 2008
|
|
1S2K
 
 | | Structure of SCP-B a member of the Eqolisin family of Peptidases in a complex with a Tripeptide Ala-Ile-His | | Descriptor: | Ala-Ile-His tripeptide, Scytalidopepsin B, TYROSINE | | Authors: | Fujinaga, M, Cherney, M.M, Oyama, H, Oda, K, James, M.N. | | Deposit date: | 2004-01-08 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The molecular structure and catalytic mechanism of a novel carboxyl peptidase from Scytalidium lignicolum
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
