8Y8K
 
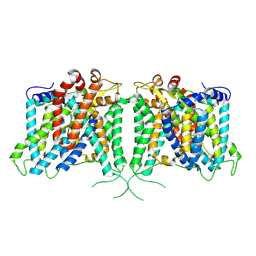 | | The structure of hAE3 | | Descriptor: | Anion exchange protein 3 | | Authors: | Jian, L, Zhang, Q, Yao, D, Wang, Q, Xia, Y, Qin, A, Cao, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | The structural insight into the functional modulation of human anion exchanger 3.
Nat Commun, 15, 2024
|
|
8ZLE
 
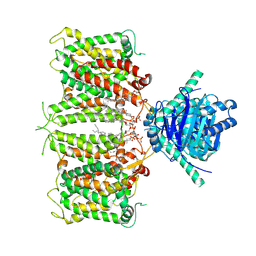 | | hAE3NTD2TMD with PT5,CLR, and Y01 | | Descriptor: | CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, [(2R)-1-octadecanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8Z)-icosa-5,8,11,14-tetraenoate, ... | | Authors: | Jian, L, Zhang, Q, Yao, D, Wang, Q, Xia, Y, Qin, A, Cao, Y. | | Deposit date: | 2024-05-19 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | The structural insight into the functional modulation of human anion exchanger 3.
Nat Commun, 15, 2024
|
|
8Y86
 
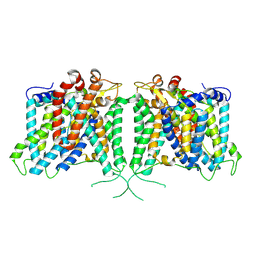 | | Human AE3 with NaHCO3- | | Descriptor: | Anion exchange protein 3, BICARBONATE ION | | Authors: | Jian, L, Zhang, Q, Yao, D, Cao, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | The structural insight into the functional modulation of human anion exchanger 3.
Nat Commun, 15, 2024
|
|
8Y85
 
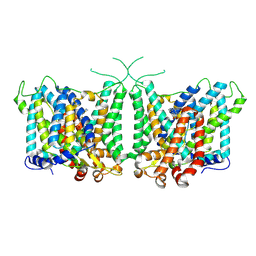 | | Human AE3 with NaHCO3- and DIDS | | Descriptor: | 2,2'-ethane-1,2-diylbis{5-[(sulfanylmethyl)amino]benzenesulfonic acid}, Anion exchange protein 3, BICARBONATE ION | | Authors: | Jian, L, Zhang, Q, Yao, D, Wang, Q, Xia, Y, Qin, A, Cao, Y. | | Deposit date: | 2024-02-05 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | The structural insight into the functional modulation of human anion exchanger 3.
Nat Commun, 15, 2024
|
|
8H9D
 
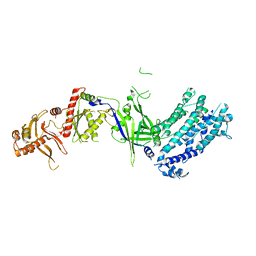 | | Crystal structure of Cas12a protein | | Descriptor: | Cas12A, MAGNESIUM ION, RNA (5'-R(P*AP*AP*UP*UP*UP*CP*UP*AP*CP*UP*AP*AP*GP*UP*GP*UP*AP*GP*AP*UP*C)-3'), ... | | Authors: | Jianwei, L, Jobichen, C, Sivaraman, J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of apo Cas12a and its complex with crRNA and DNA reveal the dynamics of ternary complex formation and target DNA cleavage.
Plos Biol., 21, 2023
|
|
6L6Q
 
 | |
3BBV
 
 | |
6LC1
 
 | |
8IOF
 
 | |
5WXP
 
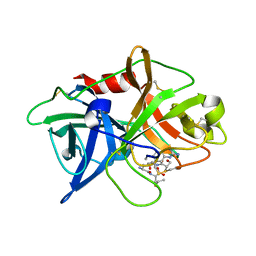 | | Crystal structure of uPA in complex with upain-2-3-W3A | | Descriptor: | ALANINE, CYSTEINE, Urokinase-type plasminogen activator chain B, ... | | Authors: | Jiang, L, Huang, M. | | Deposit date: | 2017-01-08 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cleavage of peptidic inhibitors by target protease is caused by peptide conformational transition.
Biochim. Biophys. Acta, 1862, 2018
|
|
5WXT
 
 | | Crystal structure of uPA-S195A in complex with S2444 | | Descriptor: | (2R)-N-[2-[[(2S)-1-[[4-[bis(oxidanyl)amino]phenyl]amino]-5-carbamimidamido-1-oxidanylidene-pentan-2-yl]amino]-2-oxidanylidene-ethyl]-5-oxidanylidene-pyrrolidine-2-carboxamide, Urokinase-type plasminogen activator chain B | | Authors: | Jiang, L, Huang, M. | | Deposit date: | 2017-01-08 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of uPA-S195A in complex with S2444
To Be Published
|
|
5WXQ
 
 | |
5WXF
 
 | | Crystal structure of uPA in complex with upain-2-2 | | Descriptor: | SULFATE ION, Urokinase-type plasminogen activator chain B, upain-2-2 peptide | | Authors: | Jiang, L, Huang, M. | | Deposit date: | 2017-01-07 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Cleavage of peptidic inhibitors by target protease is caused by peptide conformational transition.
Biochim. Biophys. Acta, 1862, 2018
|
|
5WXR
 
 | |
5WXO
 
 | | Crystal structure of uPA in complex with upain-2-2-W3A | | Descriptor: | Urokinase-type plasminogen activator chain B, upain-2-2-W3A peptide | | Authors: | Jiang, L, Huang, M. | | Deposit date: | 2017-01-08 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Cleavage of peptidic inhibitors by target protease is caused by peptide conformational transition.
Biochim. Biophys. Acta, 1862, 2018
|
|
5WXS
 
 | | Crystal structure of uPA in complex with S2444 | | Descriptor: | (2R)-N-[2-[[(2S)-5-carbamimidamido-1-oxidanylidene-pentan-2-yl]amino]-2-oxidanylidene-ethyl]-5-oxidanylidene-1,2-dihydropyrrole-2-carboxamide, Urokinase-type plasminogen activator chain B | | Authors: | Jiang, L, Huang, M. | | Deposit date: | 2017-01-08 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of uPA in complex with S2444
To Be Published
|
|
3BBX
 
 | |
3BBU
 
 | |
4X0W
 
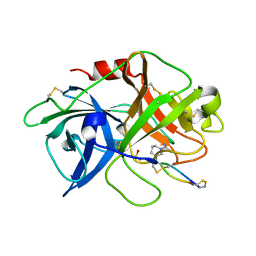 | | The crystal structure of mupain-1-17 in complex with murinised human uPA | | Descriptor: | SULFATE ION, Urokinase-type plasminogen activator, mupain-1-17, ... | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-24 | | Release date: | 2015-10-21 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinctive binding modes and inhibitory mechanisms of two peptidic inhibitors of urokinase-type plasminogen activator with isomeric P1 residues.
Int.J.Biochem.Cell Biol., 62, 2015
|
|
4X1P
 
 | | The crystal structure of mupain-1-17 in complex with murinised human uPA at pH4.6 | | Descriptor: | MUPAIN-1-17, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-25 | | Release date: | 2015-10-21 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Distinctive binding modes and inhibitory mechanisms of two peptidic inhibitors of urokinase-type plasminogen activator with isomeric P1 residues.
Int.J.Biochem.Cell Biol., 62, 2015
|
|
3M61
 
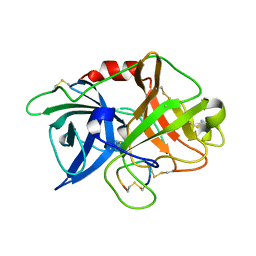 | | Crystal structure of complex of urokinase and a upain-1 variant(W3A) in pH4.6 condition | | Descriptor: | Urokinase-type plasminogen activator, upain-1 W3A | | Authors: | Jiang, L, Yuan, C, Wind, T, Andreasen, P.A, Chen, L, Meehan, E.J, Huang, M. | | Deposit date: | 2010-03-15 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of complex of urokinase and a upain-1 variant(W3A) in pH4.6 condition
TO BE PUBLISHED
|
|
2TOB
 
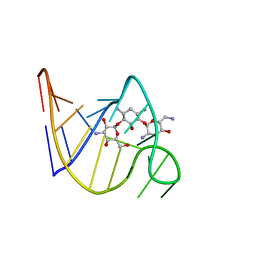 | | SOLUTION STRUCTURE OF THE TOBRAMYCIN-RNA APTAMER COMPLEX, NMR, 13 STRUCTURES | | Descriptor: | 1,3-DIAMINO-4,5,6-TRIHYDROXY-CYCLOHEXANE, 2,6-diammonio-2,3,6-trideoxy-alpha-D-glucopyranose, 3-ammonio-3-deoxy-alpha-D-glucopyranose, ... | | Authors: | Jiang, L, Patel, D.J. | | Deposit date: | 1998-06-17 | | Release date: | 1999-06-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tobramycin-RNA aptamer complex.
Nat.Struct.Biol., 5, 1998
|
|
1NEM
 
 | | Saccharide-RNA recognition in the neomycin B / RNA aptamer complex | | Descriptor: | 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranose, 2,6-diamino-2,6-dideoxy-beta-L-idopyranose-(1-3)-beta-D-ribofuranose, 2-DEOXY-D-STREPTAMINE, ... | | Authors: | Jiang, L, Majumdar, A, Hu, W, Jaishree, T.J, Xu, W, Patel, D.J. | | Deposit date: | 1999-03-15 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Saccharide-RNA recognition in a complex formed between neomycin B and an RNA aptamer
Structure Fold.Des., 7, 1999
|
|
4DVB
 
 | | The crystal structure of the Fab fragment of pro-uPA antibody mAb-112 | | Descriptor: | Fab fragment of pro-uPA antibody mAb-112, SULFATE ION, TETRAETHYLENE GLYCOL | | Authors: | Jiang, L, Botkjaer, K.A, Andersen, L.M, Yuan, C, Andreasen, P.A, Huang, M. | | Deposit date: | 2012-02-23 | | Release date: | 2013-01-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Rezymogenation of active urokinase induced by an inhibitory antibody.
Biochem.J., 449, 2013
|
|
1TOB
 
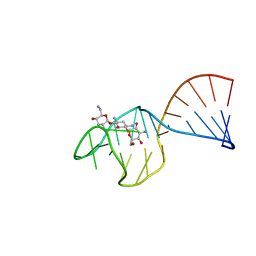 | | SACCHARIDE-RNA RECOGNITION IN AN AMINOGLYCOSIDE ANTIBIOTIC-RNA APTAMER COMPLEX, NMR, 7 STRUCTURES | | Descriptor: | 1,3-DIAMINO-5,6-DIHYDROXYCYCLOHEXANE, 2,6-diammonio-2,3,6-trideoxy-alpha-D-glucopyranose, 3-ammonio-3-deoxy-alpha-D-glucopyranose, ... | | Authors: | Jiang, L, Suri, A.K, Fiala, R, Patel, D.J. | | Deposit date: | 1996-12-12 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Saccharide-RNA recognition in an aminoglycoside antibiotic-RNA aptamer complex.
Chem.Biol., 4, 1997
|
|
