7Z8O
 
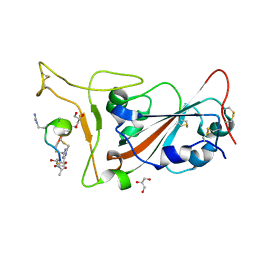 | | Crystal structure of SARS-CoV-2 S RBD in complex with a stapled peptide | | Descriptor: | 2,4,6-tris(chloromethyl)-1,3,5-triazine, GLYCEROL, Spike protein S1, ... | | Authors: | Brear, P, Chen, L, Gaynor, K, Harman, M, Dods, R, Hyvonen, M. | | Deposit date: | 2022-03-18 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Multivalent bicyclic peptides are an effective antiviral modality that can potently inhibit SARS-CoV-2.
Nat Commun, 14, 2023
|
|
6VT2
 
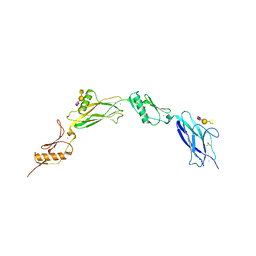 | |
6VU6
 
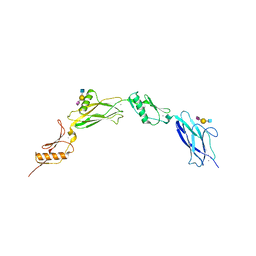 | |
6VS7
 
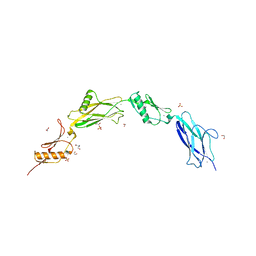 | | Sialic acid binding region of Streptococcus Sanguinis SK1 adhesin | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Adhesin, ... | | Authors: | Stubbs, H.E, Iverson, T.M. | | Deposit date: | 2020-02-10 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tandem sialoglycan-binding modules in a Streptococcus sanguinis serine-rich repeat adhesin create target dependent avidity effects.
J.Biol.Chem., 295, 2020
|
|
6BR4
 
 | | Crystal structure of Escherichia coli DsbA in complex with {N}-methyl-1-(3-thiophen-2-ylphenyl)methanamine | | Descriptor: | COPPER (II) ION, Thiol:disulfide interchange protein DsbA, ~{N}-methyl-1-(3-thiophen-2-ylphenyl)methanamine | | Authors: | Heras, B, Totsika, M, Paxman, J.J, Wang, G, Scanlon, M.J, Martin, J.L. | | Deposit date: | 2017-11-29 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Inhibition of Diverse DsbA Enzymes in Multi-DsbA Encoding Pathogens.
Antioxid. Redox Signal., 29, 2018
|
|
6IVV
 
 | | Structure of peptidyl-tRNA hydrolase from Acinetobacter baumannii with multiple surface binding regions at 1.26A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Viswanathan, V, Sharma, P, Chaudhary, A, Sharma, S, Singh, T.P. | | Deposit date: | 2018-12-04 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structure of peptide t-RNA hydrolase from Acinetobacter baumannii with multiple surface binding sites at 1.26 Angstrom resolution.
To Be Published
|
|
6IX6
 
 | | Crystal structure of the complex of peptidyl-tRNA hydrolase with N-propanol at 1.43 A resolution | | Descriptor: | N-PROPANOL, Peptidyl-tRNA hydrolase | | Authors: | Viswanathan, V, Sharma, P, Chaudhary, A, Sharma, S, Singh, T.P. | | Deposit date: | 2018-12-09 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of the complex of peptidyl-tRNA hydrolase with N-propanol at 1.43 A resolution
To Be Published
|
|
6IYE
 
 | |
7VQI
 
 | |
7Y3U
 
 | | Crystal structure of the complex of Lactoperoxidase with Nitric oxide at 2.50A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Singh, P.K, Viswanathan, V, Ahmad, N, Rani, C, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-06-13 | | Release date: | 2022-06-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the complex of Lactoperoxidase with Nitric oxide at 2.50A resolution
To Be Published
|
|
9C10
 
 | | AMG 193, a clinical stage MTA-cooperative PRMT5 inhibitor, drives anti-tumor activity preclinically and in patients with MTAP-deleted cancers | | Descriptor: | (4-amino-1,3-dihydrofuro[3,4-c][1,7]naphthyridin-8-yl){(3S)-3-[4-(trifluoromethyl)phenyl]morpholin-4-yl}methanone, 5'-DEOXY-5'-METHYLTHIOADENOSINE, DIMETHYL SULFOXIDE, ... | | Authors: | Ghimire-Rijal, S, Mukund, S. | | Deposit date: | 2024-05-28 | | Release date: | 2024-10-02 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | AMG 193, a Clinical Stage MTA-Cooperative PRMT5 Inhibitor, Drives Antitumor Activity Preclinically and in Patients with MTAP-Deleted Cancers.
Cancer Discov, 15, 2025
|
|
6O5Z
 
 | | Crystal Structure of the human MLKL pseudokinase domain bound to compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-fluoranyl-5-(trifluoromethyl)phenyl]-3-[4-[methyl-[2-[(3-sulfamoylphenyl)amino]pyrimidin-4-yl]amino]phenyl]urea, Mixed lineage kinase domain-like protein | | Authors: | Cowan, A.D, Murphy, J.M, Pierotti, C.L, Lessene, G.L, Czabotar, P.E. | | Deposit date: | 2019-03-04 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Potent Inhibition of Necroptosis by Simultaneously Targeting Multiple Effectors of the Pathway.
Acs Chem.Biol., 15, 2020
|
|
6AP8
 
 | |
6AP6
 
 | | Crystal Structure of DAD2 in complex with tolfenamic acid | | Descriptor: | 2-[(3-chloro-2-methylphenyl)amino]benzoic acid, Probable strigolactone esterase DAD2 | | Authors: | Hamiaux, C. | | Deposit date: | 2017-08-17 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibition of strigolactone receptors byN-phenylanthranilic acid derivatives: Structural and functional insights.
J. Biol. Chem., 293, 2018
|
|
6AP7
 
 | |
7VQH
 
 | |
4WET
 
 | | Crystal structure of E.Coli DsbA in complex with compound 16 | | Descriptor: | 1,2-ETHANEDIOL, N-({4-methyl-2-[4-(trifluoromethyl)phenyl]-1,3-thiazol-5-yl}carbonyl)-L-tyrosine, SODIUM ION, ... | | Authors: | Ilyichova, O.V, Scanlon, M.J. | | Deposit date: | 2014-09-11 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Application of Fragment-Based Screening to the Design of Inhibitors of Escherichia coli DsbA.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7TBA
 
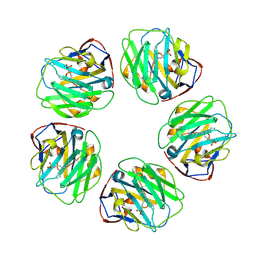 | | Pentraxin - ligand complex | | Descriptor: | C-reactive protein, CALCIUM ION, [3-(dibutylamino)propyl]phosphonic acid | | Authors: | Shing, K.S.C.T, Morton, C.J, Parker, M.W. | | Deposit date: | 2021-12-21 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A novel phosphocholine-mimetic inhibits a pro-inflammatory conformational change in C-reactive protein.
Embo Mol Med, 15, 2023
|
|
3H93
 
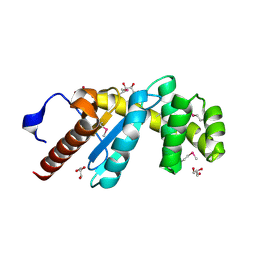 | | Crystal Structure of Pseudomonas aeruginosa DsbA | | Descriptor: | GLYCEROL, Thiol:disulfide interchange protein dsbA | | Authors: | Shouldice, S.R. | | Deposit date: | 2009-04-29 | | Release date: | 2009-12-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Characterization of the DsbA Oxidative Folding Catalyst from Pseudomonas aeruginosa Reveals a Highly Oxidizing Protein that Binds Small Molecules.
Antioxid Redox Signal, 12, 2010
|
|
6SS4
 
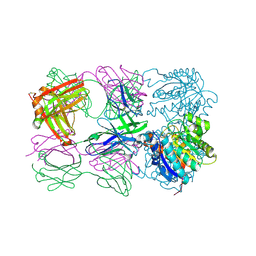 | | Structure of arginase-2 in complex with the inhibitory human antigen-binding fragment Fab C0021181 | | Descriptor: | Arginase-2, mitochondrial, Fab C0021181 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6SS2
 
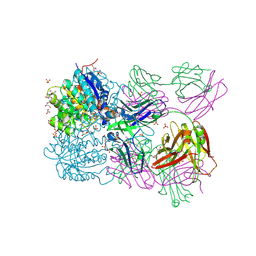 | | Structure of arginase-2 in complex with the inhibitory human antigen-binding fragment Fab C0021158 | | Descriptor: | Arginase-2, mitochondrial, Fab C0021158 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
9N4Z
 
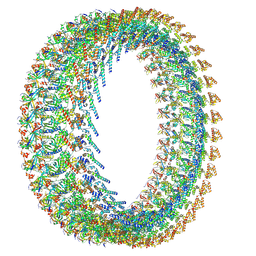 | | CCW Flagellar Switch Complex - FliF, FliG, FliM, and FliN forming 34-mer C-ring from Salmonella | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Singh, P.K, Iverson, T.M. | | Deposit date: | 2025-02-03 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Improving CryoEM maps of symmetry-mismatched macromolecular assemblies: A case study on the flagellar motor.
J.Struct.Biol., 217, 2025
|
|
9N49
 
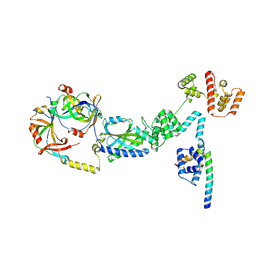 | | C-ring - single subunit of the 34-mer CCW flagellar switch complex - FliF, FliG, FliM, and FliN from Salmonella | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Singh, P.K, Iverson, T.M. | | Deposit date: | 2025-02-02 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Improving CryoEM maps of symmetry-mismatched macromolecular assemblies: A case study on the flagellar motor.
J.Struct.Biol., 217, 2025
|
|
8OYT
 
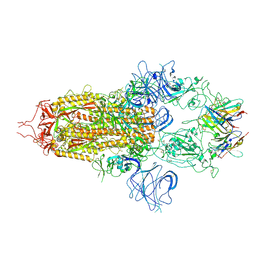 | | Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '3 up' RBD conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H6 nanobody, ... | | Authors: | Weckener, M, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
8OWT
 
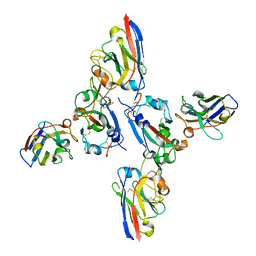 | | SARS-CoV-2 spike RBD with A8 and H3 nanobodies bound | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody A8, ... | | Authors: | Mikolajek, H, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
