7DZF
 
 | | Intermediate of FABP with a delay time of 10 ns | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZG
 
 | | Intermediate of FABP with a delay time of 30 ns | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZI
 
 | | intermediate of FABP with a delay time of 300 ns | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZJ
 
 | | Fabp protein before hv | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZK
 
 | | Fabp protein after hv | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZH
 
 | | intermediate of FABP with a delay time of 100 ns | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZL
 
 | | A69C-M71L mutant of Fabp protein | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7FEO
 
 | | Crystal structure of AtMBD5 MBD domain | | Descriptor: | Methyl-CpG-binding domain-containing protein 5, SULFATE ION | | Authors: | Zhou, M.Q, Wu, Z.B, Liu, K, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-21 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Family-wide Characterization of Methylated DNA Binding Ability of Arabidopsis MBDs.
J.Mol.Biol., 434, 2022
|
|
7FEF
 
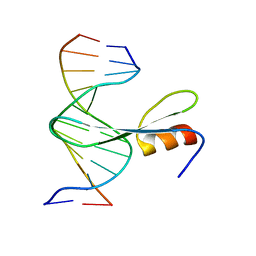 | | Crystal structure of AtMBD6 with DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain-containing protein 6 | | Authors: | Wu, Z.B, Liu, K, Min, J.R. | | Deposit date: | 2021-07-19 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Family-wide Characterization of Methylated DNA Binding Ability of Arabidopsis MBDs.
J.Mol.Biol., 434, 2022
|
|
5JKG
 
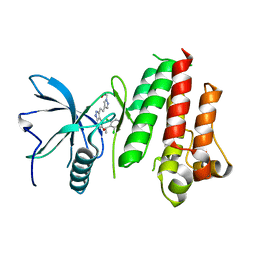 | | The crystal structure of FGFR4 kinase domain in complex with LY2874455 | | Descriptor: | 2-[4-[E-2-[5-[(1R)-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-1H-indazol-3-yl]ethenyl]pyrazol-1-yl]ethanol, Fibroblast growth factor receptor 4 | | Authors: | Wu, D, Chen, L, Chen, Y. | | Deposit date: | 2016-04-26 | | Release date: | 2016-10-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Crystal Structure of the FGFR4/LY2874455 Complex Reveals Insights into the Pan-FGFR Selectivity of LY2874455
Plos One, 11, 2016
|
|
8CEG
 
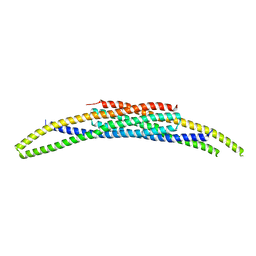 | | BAR domain protein FAM92A1 essential for mitochondrial membrane remodeling | | Descriptor: | CBY1-interacting BAR domain-containing protein 1 | | Authors: | Kajander, T, Fudo, S, Yan, Z, Zhao, H. | | Deposit date: | 2023-02-01 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Membrane remodeling by FAM92A1 during brain development regulates neuronal morphology, synaptic function, and cognition.
Nat Commun, 15, 2024
|
|
4DSB
 
 | | Complex Structure of Abscisic Acid Receptor PYL3 with (+)-ABA in Spacegroup of I 212121 at 2.70A | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3 | | Authors: | Zhang, X, Zhang, Q, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
9BRZ
 
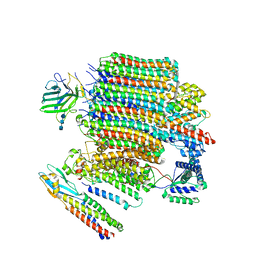 | | V0-only V-ATPase and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-12 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRU
 
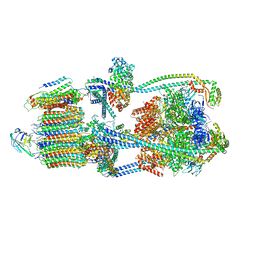 | | Intact V-ATPase State 1 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRT
 
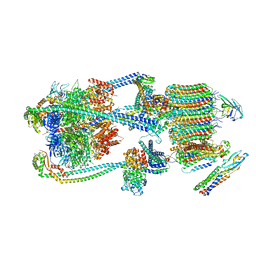 | | Intact V-ATPase State 1 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRA
 
 | | Intact V-ATPase State 2 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRR
 
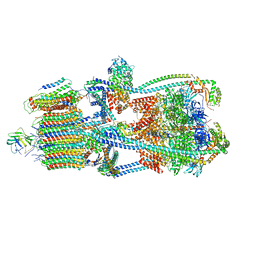 | | Intact V-ATPase State 3 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRS
 
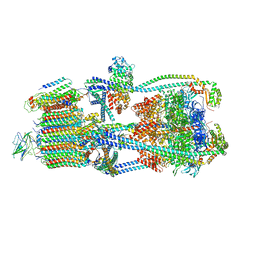 | | Intact V-ATPase State 2 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRY
 
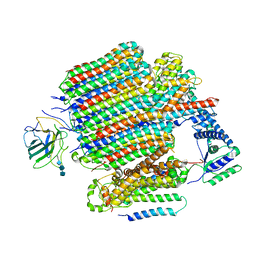 | | V0-only V-ATPase in synaptophysin gene knock-out mouse brain isolated synaptic vesicles | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-12 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRQ
 
 | | Intact V-ATPase State 3 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
5XFJ
 
 | | Crystal structure of LY2874455 in complex of FGFR4 gatekeeper mutation (V550M) | | Descriptor: | 2-[4-[E-2-[5-[(1R)-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-1H-indazol-3-yl]ethenyl]pyrazol-1-yl]ethanol, Fibroblast growth factor receptor 4 | | Authors: | Wu, D, Chen, Y. | | Deposit date: | 2017-04-10 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | LY2874455 potently inhibits FGFR gatekeeper mutants and overcomes mutation-based resistance.
Chem. Commun. (Camb.), 54, 2018
|
|
5XFF
 
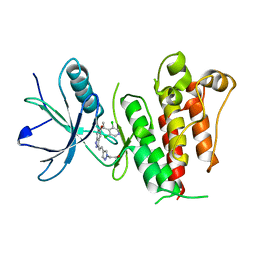 | | Crystal structure of LY2874455 in complex of FGFR4 gatekeeper mutation (V550L) | | Descriptor: | 2-[4-[E-2-[5-[(1R)-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-1H-indazol-3-yl]ethenyl]pyrazol-1-yl]ethanol, Fibroblast growth factor receptor 4 | | Authors: | Wu, D, Chen, Y. | | Deposit date: | 2017-04-10 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | LY2874455 potently inhibits FGFR gatekeeper mutants and overcomes mutation-based resistance.
Chem. Commun. (Camb.), 54, 2018
|
|
4QNP
 
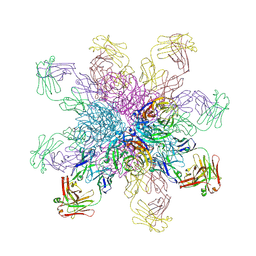 | | Crystal structure of the 2009 pandemic H1N1 influenza virus neuraminidase with a neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wan, H.Q, Yang, H, Shore, D.A, Garten, R.J, Couzens, L, Gao, J, Jiang, L.L, Carney, P.J, Villanueva, J, Stevens, J, Eichelberger, M.C. | | Deposit date: | 2014-06-18 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural characterization of a protective epitope spanning A(H1N1)pdm09 influenza virus neuraminidase monomers.
Nat Commun, 6, 2015
|
|
4E0L
 
 | | FYLLYYT segment from human Beta 2 Microglobulin (62-68) displayed on 54-membered macrocycle scaffold | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cyclic pseudo-peptide FYLLYYT(ORN)KN(HAO)SA(ORN) | | Authors: | Zhao, M, Liu, C, Michael, S.R, Eisenberg, D. | | Deposit date: | 2012-03-04 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Out-of-register beta-sheets suggest a pathway to toxic amyloid aggregates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6LT0
 
 | | cryo-EM structure of C9ORF72-SMCR8-WDR41 | | Descriptor: | Guanine nucleotide exchange C9orf72, Guanine nucleotide exchange protein SMCR8, WD repeat-containing protein 41 | | Authors: | Tang, D, Sheng, J, Xu, L, Zhan, X, Yan, C, Qi, S. | | Deposit date: | 2020-01-21 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of C9ORF72-SMCR8-WDR41 reveals the role as a GAP for Rab8a and Rab11a.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
