2PPL
 
 | | Human Pancreatic lipase-related protein 1 | | Descriptor: | CALCIUM ION, Pancreatic lipase-related protein 1, SODIUM ION | | Authors: | Walker, J.R, Davis, T, Seitova, A, Butler-Cole, C, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-30 | | Release date: | 2007-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Human Pancreatic Lipase-related Protein 1.
To be Published
|
|
2PBC
 
 | | FK506-binding protein 2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FK506-binding protein 2 | | Authors: | Walker, J.R, Neculai, D, Davis, T, Butler-Cole, C, Sicheri, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-28 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of FK506-Binding Protein 2
To be Published
|
|
2P0R
 
 | | Structure of Human Calpain 9 in complex with Leupeptin | | Descriptor: | CALCIUM ION, Calpain-9, leupeptin | | Authors: | Davis, T.L, Paramanathan, R, Walker, J.R, Butler-Cole, C, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-01 | | Release date: | 2007-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Human Minicalpains bound to Inhibitors
To be Published
|
|
2NQA
 
 | | Catalytic Domain of Human Calpain 8 | | Descriptor: | CALCIUM ION, Calpain-8, Leupeptin Inhibitor | | Authors: | Davis, T.L, Paramanathan, R, Butler-Cole, C, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-10-30 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Human Calpain 8
To be Published
|
|
2Q2F
 
 | | Structure of the human Selenoprotein S (VCP-interacting membrane protein) | | Descriptor: | CHLORIDE ION, GLYCEROL, Selenoprotein S | | Authors: | Walker, J.R, Paramanathan, R, Butler-Cole, C, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-28 | | Release date: | 2007-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Human Selenoprotein S (VCP-interacting membrane protein).
To be Published
|
|
2QOB
 
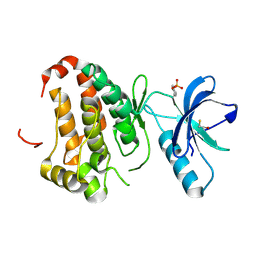 | | Human EphA3 kinase domain, base structure | | Descriptor: | BETA-MERCAPTOETHANOL, Ephrin receptor | | Authors: | Davis, T, Walker, J.R, Newman, E.M, Mackenzie, F, Butler-Cole, C, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-20 | | Release date: | 2007-08-21 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Autoregulation by the Juxtamembrane Region of the Human Ephrin Receptor Tyrosine Kinase A3 (EphA3).
Structure, 16, 2008
|
|
2QOO
 
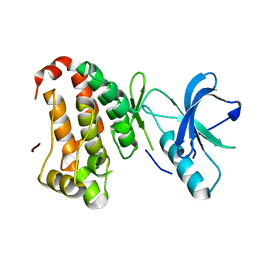 | | Human EphA3 kinase and juxtamembrane region, Y596F:Y602F:Y742F triple mutant | | Descriptor: | Ephrin receptor | | Authors: | Davis, T, Walker, J.R, Newman, E.M, Mackenzie, F, Butler-Cole, C, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-20 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Autoregulation by the Juxtamembrane Region of the Human Ephrin Receptor Tyrosine Kinase A3 (EphA3).
Structure, 16, 2008
|
|
2QOF
 
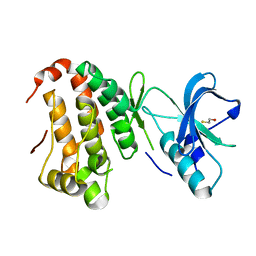 | | Human EphA3 kinase and juxtamembrane region, Y596F mutant | | Descriptor: | BETA-MERCAPTOETHANOL, Ephrin receptor | | Authors: | Davis, T, Walker, J.R, Newman, E.M, Mackenzie, F, Butler-Cole, C, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-20 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Autoregulation by the Juxtamembrane Region of the Human Ephrin Receptor Tyrosine Kinase A3 (EphA3).
Structure, 16, 2008
|
|
2QO2
 
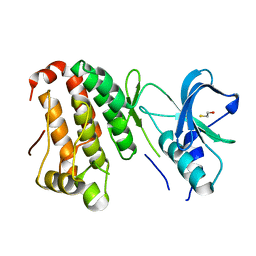 | | Human EphA3 kinase and juxtamembrane region, dephosphorylated, apo structure | | Descriptor: | BETA-MERCAPTOETHANOL, Ephrin receptor | | Authors: | Davis, T, Walker, J.R, Newman, E.M, Mackenzie, F, Butler-Cole, C, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-19 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Autoregulation by the Juxtamembrane Region of the Human Ephrin Receptor Tyrosine Kinase A3 (EphA3).
Structure, 16, 2008
|
|
2R2P
 
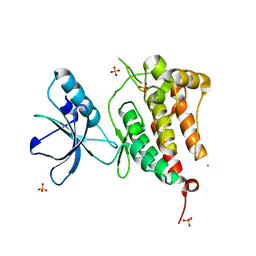 | | Kinase domain of human ephrin type-A receptor 5 (EphA5) | | Descriptor: | CHLORIDE ION, Ephrin type-A receptor 5, SULFATE ION | | Authors: | Walker, J.R, Cuerrier, D, Butler-Cole, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-08-27 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinase Domain of Human Ephrin Type-A Receptor 5 (EphA5).
To be Published
|
|
4F7B
 
 | | Structure of the lysosomal domain of limp-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Neculai, D, Ravichandran, M, Seitova, A, Neculai, M, Pizzaro, J.C, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Dhe-Paganon, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-05-15 | | Release date: | 2013-10-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of LIMP-2 provides functional insights with implications for SR-BI and CD36.
Nature, 504, 2013
|
|
3T6J
 
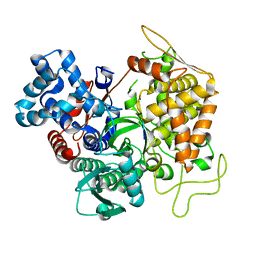 | |
4ZC9
 
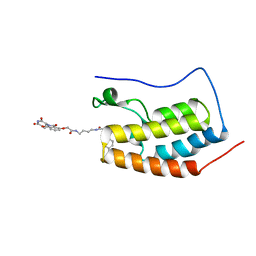 | | Crystal Structure of the BRD4a/DB-2-190 complex | | Descriptor: | 2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]-N-(4-{[({2-[(3S)-2,6-dioxopiperidin-3-yl]-1,3-dioxo-2,3-dihydro-1H-isoindol-4-yl}oxy)acetyl]amino}butyl)acetamide, Bromodomain-containing protein 4 | | Authors: | Seo, H.-S, DeAngelo, S, Bradner, J.E. | | Deposit date: | 2015-04-15 | | Release date: | 2015-11-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation.
Science, 348, 2015
|
|
4I6L
 
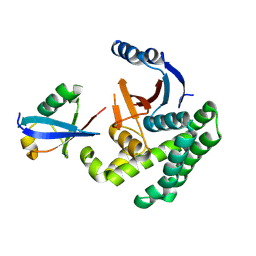 | |
8THO
 
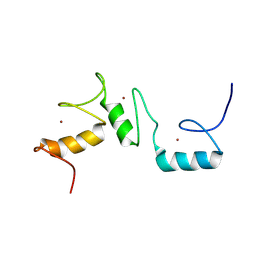 | | Solution structure of the zinc finger repeat domain of BCL11A (ZnF456) | | Descriptor: | B-cell lymphoma/leukemia 11A, ZINC ION | | Authors: | Viennet, T, Yin, M, Orkin, S.H, Arthanari, H. | | Deposit date: | 2023-07-17 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-05 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the DNA-binding mechanism of BCL11A: The integral role of ZnF6.
Structure, 32, 2024
|
|
4EBB
 
 | | Structure of DPP2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 2, ZINC ION | | Authors: | Shewchuk, L.M, Hassell, A.H, Sweitzer, S.M, Sweitzer, T.D, McDevitt, P.J, Kennedy-Wilson, K.M, Johanson, K.O. | | Deposit date: | 2012-03-23 | | Release date: | 2012-09-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Human DPP7 Reveal the Molecular Basis of Specific Inhibition and the Architectural Diversity of Proline-Specific Peptidases.
Plos One, 7, 2012
|
|
4ORH
 
 | | Crystal structure of RNF8 bound to the UBC13/MMS2 heterodimer | | Descriptor: | E3 ubiquitin-protein ligase RNF8, Ubiquitin-conjugating enzyme E2 N, Ubiquitin-conjugating enzyme E2 variant 2, ... | | Authors: | Campbell, S.J, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2014-02-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.802 Å) | | Cite: | Molecular insights into the function of RING finger (RNF)-containing proteins hRNF8 and hRNF168 in Ubc13/Mms2-dependent ubiquitylation.
J.Biol.Chem., 287, 2012
|
|
3RZ3
 
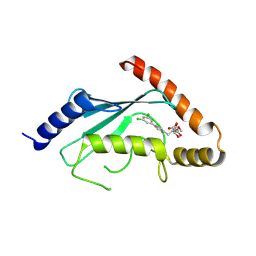 | | Human Cdc34 E2 in complex with CC0651 inhibitor | | Descriptor: | 4,5-dideoxy-5-(3',5'-dichlorobiphenyl-4-yl)-4-[(methoxyacetyl)amino]-L-arabinonic acid, Ubiquitin-conjugating enzyme E2 R1 | | Authors: | Ceccarelli, D.F, Webb, D.R, Sicheri, F. | | Deposit date: | 2011-05-11 | | Release date: | 2011-07-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An allosteric inhibitor of the human cdc34 ubiquitin conjugating enzyme
Cell(Cambridge,Mass.), 145, 2011
|
|
3T6B
 
 | |
1PZL
 
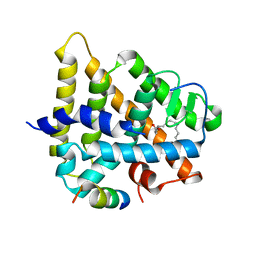 | | Crystal structure of HNF4a LBD in complex with the ligand and the coactivator SRC-1 peptide | | Descriptor: | Hepatocyte nuclear factor 4-alpha, MYRISTIC ACID, steroid receptor coactivator-1 | | Authors: | Duda, K, Chi, Y.-I, Dhe-paganon, S, Shoelson, S. | | Deposit date: | 2003-07-11 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for HNF-4alpha Activation by Ligand and Coactivator Binding
J.Biol.Chem., 279, 2004
|
|
7JIF
 
 | | HRAS A59T GppNHp | | Descriptor: | GLYCEROL, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Johnson, C.W, Haigis, K.M. | | Deposit date: | 2020-07-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.757 Å) | | Cite: | Regulation of GTPase function by autophosphorylation.
Mol.Cell, 82, 2022
|
|
7JIH
 
 | | HRAS A59E GppNHp | | Descriptor: | GLYCEROL, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Johnson, C.W, Haigis, K.M. | | Deposit date: | 2020-07-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | Regulation of GTPase function by autophosphorylation.
Mol.Cell, 82, 2022
|
|
7JIG
 
 | | HRAS A59T GppNHp crystal 2 | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Johnson, C.W, Haigis, K.M. | | Deposit date: | 2020-07-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.322 Å) | | Cite: | Regulation of GTPase function by autophosphorylation.
Mol.Cell, 82, 2022
|
|
7JII
 
 | | HRAS A59E GDP | | Descriptor: | CALCIUM ION, GTPase HRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Johnson, C.W, Haigis, K.M. | | Deposit date: | 2020-07-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Regulation of GTPase function by autophosphorylation.
Mol.Cell, 82, 2022
|
|
3K9P
 
 | | The crystal structure of E2-25K and ubiquitin complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 K | | Authors: | Kang, G.B, Ko, S, Song, S.M, Lee, W, Eom, S.H. | | Deposit date: | 2009-10-16 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of E2-25K/UBB+1 interaction leading to proteasome inhibition and neurotoxicity
J.Biol.Chem., 285, 2010
|
|
