8A3H
 
 | | Cellobiose-derived imidazole complex of the endoglucanase cel5A from Bacillus agaradhaerens at 0.97 A resolution | | Descriptor: | (5R,6R,7R,8S)-7,8-dihydroxy-5-(hydroxymethyl)-5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-6-yl beta-D-glucopyranoside, ACETATE ION, GLYCEROL, ... | | Authors: | Varrot, A, Schulein, M, Pipelier, M, Vasella, A, Davies, G.J. | | Deposit date: | 1999-01-20 | | Release date: | 2000-01-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Lateral Protonation of a Glycosidase Inhibitor. Structure of the Bacillus agaradhaerens Cel5A in Complex with a Cellobiose-Derived Imidazole at 0.97 A Resolution
J.Am.Chem.Soc., 121, 1999
|
|
6ZFA
 
 | | Structure of the catalytic domain of human endo-alpha-mannosidase MANEA in complex with GlcIFG, alpha-1,2-mannobiose and hexatungstotellurate(VI) TEW | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, 6-tungstotellurate(VI), ... | | Authors: | Sobala, L.F, Fernandes, P.Z, Hakki, Z, Thompson, A.J, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-16 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZFN
 
 | | Structure of an inactive E404Q variant of the catalytic domain of human endo-alpha-mannosidase MANEA in complex with 1-methyl alpha-1,2-mannobiose | | Descriptor: | Glycoprotein endo-alpha-1,2-mannosidase, SULFATE ION, alpha-D-mannopyranose-(1-2)-methyl alpha-D-mannopyranoside | | Authors: | Sobala, L.F, Fernandes, P.Z, Hakki, Z, Thompson, A.J, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZDC
 
 | | Structure of the catalytic domain of human endo-alpha-mannosidase MANEA in complex with nickel | | Descriptor: | Glycoprotein endo-alpha-1,2-mannosidase, NICKEL (II) ION | | Authors: | Sobala, L.F, Fernandes, P.Z, Hakki, Z, Thompson, A.J, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZFQ
 
 | | Structure of the catalytic domain of human endo-alpha-mannosidase MANEA in complex with bis-tris | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycoprotein endo-alpha-1,2-mannosidase | | Authors: | Sobala, L.F, Fernandes, P.Z, Hakki, Z, Thompson, A.J, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZDF
 
 | | Structure of the catalytic domain of human endo-alpha-mannosidase MANEA in complex with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Glycoprotein endo-alpha-1,2-mannosidase | | Authors: | Sobala, L.F, Fernandes, P.Z, Hakki, Z, Thompson, A.J, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZDK
 
 | | Structure of the catalytic domain of human endo-alpha-mannosidase MANEA in complex with HEPES and hexatungstotellurate(VI) TEW | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 6-tungstotellurate(VI), Glycoprotein endo-alpha-1,2-mannosidase, ... | | Authors: | Sobala, L.F, Fernandes, P.Z, Hakki, Z, Thompson, A.J, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZJ6
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with cyclohexylmethyl-Glc-1,3-isofagomine | | Descriptor: | 1,2-ETHANEDIOL, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, ACETATE ION, ... | | Authors: | Thompson, A.J, Sobala, L.F, Fernandes, P.Z, Hakki, Z, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-27 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5BY3
 
 | | A novel family GH115 4-O-Methyl-alpha-glucuronidase, BtGH115A, with specificity for decorated arabinogalactans | | Descriptor: | BtGH115A, NICKEL (II) ION, SULFATE ION | | Authors: | Lammerts van Bueren, A, Davies, G.J, Turkenburg, J.P. | | Deposit date: | 2015-06-10 | | Release date: | 2015-07-22 | | Last modified: | 2015-12-09 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural and Functional Characterization of a Novel Family GH115 4-O-Methyl-alpha-Glucuronidase with Specificity for Decorated Arabinogalactans.
J.Mol.Biol., 427, 2015
|
|
5E9B
 
 | | Crystal structure of human heparanase in complex with HepMer M09S05a | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, CHLORIDE ION, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2015-10-14 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural characterization of human heparanase reveals insights into substrate recognition.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5E98
 
 | |
5E9C
 
 | | Crystal structure of human heparanase in complex with heparin tetrasaccharide dp4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2015-10-15 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural characterization of human heparanase reveals insights into substrate recognition.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5E8M
 
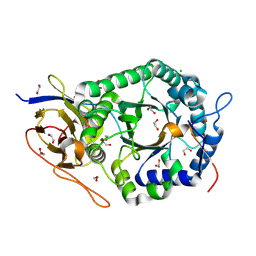 | | Crystal structure of human heparanase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2015-10-14 | | Release date: | 2015-11-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural characterization of human heparanase reveals insights into substrate recognition.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5E97
 
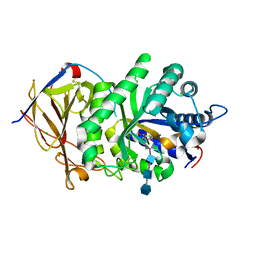 | | Glycoside Hydrolase ligand structure 1 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2015-10-14 | | Release date: | 2015-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural characterization of human heparanase reveals insights into substrate recognition.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4JFS
 
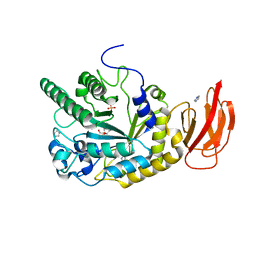 | | Crystal structure of a bacterial fucosidase with iminosugar inhibitor 4-epi-(+)-Codonopsinine | | Descriptor: | (2S,3S,4R,5S)-2-(4-methoxyphenyl)-1,5-dimethylpyrrolidine-3,4-diol, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Wright, D.W, Davies, G.J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | alpha-L-fucosidase inhibition by pyrrolidine-ferrocene hybrids: rationalization of ligand-binding properties by structural studies.
Chemistry, 19, 2013
|
|
4JL1
 
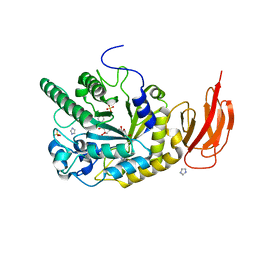 | | Crystal structure of a bacterial fucosidase with a multivalent iminocyclitol inhibitor | | Descriptor: | (3S,4R,5S)-N-benzyl-3,4-dihydroxy-5-methyl-D-prolinamide, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wright, D.W, Davies, G.J. | | Deposit date: | 2013-03-12 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Exploring a Multivalent Approach to alpha-L-Fucosidase Inhibition
EUR.J.ORG.CHEM., 2013, 2013
|
|
4JFU
 
 | | Crystal structure of a bacterial fucosidase with iminosugar inhibitor | | Descriptor: | (2S,3R,4S,5S)-2-methyl-5-(4-methylphenyl)pyrrolidine-3,4-diol, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wright, D.W, Davies, G.J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | alpha-L-fucosidase inhibition by pyrrolidine-ferrocene hybrids: rationalization of ligand-binding properties by structural studies.
Chemistry, 19, 2013
|
|
4JFV
 
 | | Crystal structure of a bacterial fucosidase with iminosugar inhibitor (2S,3S,4R,5S)-2-[N-(methylferrocene)]aminoethyl-5-methylpyrrolidine-3,4-diol | | Descriptor: | (3alpha)-[({2-[(2S,3S,4R,5S)-3,4-dihydroxy-5-methylpyrrolidin-2-yl]ethyl}amino)methyl]ferrocene, IMIDAZOLE, SULFATE ION, ... | | Authors: | Wright, D.W, Davies, G.J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | alpha-L-fucosidase inhibition by pyrrolidine-ferrocene hybrids: rationalization of ligand-binding properties by structural studies.
Chemistry, 19, 2013
|
|
4JFW
 
 | | Crystal structure of a bacterial fucosidase with iminosugar inhibitor (2S,3S,4R,5S)-2-[N-(propylferrocene)]aminoethyl-5-methylpyrrolidine-3,4-diol | | Descriptor: | (3alpha)-[3-({2-[(2S,3S,4R,5S)-3,4-dihydroxy-5-methylpyrrolidin-2-yl]ethyl}amino)propyl]ferrocene, IMIDAZOLE, SULFATE ION, ... | | Authors: | Wright, D.W, Davies, G.J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | alpha-L-fucosidase inhibition by pyrrolidine-ferrocene hybrids: rationalization of ligand-binding properties by structural studies.
Chemistry, 19, 2013
|
|
4JFT
 
 | | Crystal structure of a bacterial fucosidase with iminosugar inhibitor N-desmethyl-4-epi-(+)-Codonopsinine | | Descriptor: | (2S,3S,4R,5S)-2-(4-methoxyphenyl)-5-methylpyrrolidine-3,4-diol, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wright, D.W, Davies, G.J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | alpha-L-fucosidase inhibition by pyrrolidine-ferrocene hybrids: rationalization of ligand-binding properties by structural studies.
Chemistry, 19, 2013
|
|
4JL2
 
 | | Crystal structure of a bacterial fucosidase with a monovalent iminocyclitol inhibitor | | Descriptor: | (3S,4R,5S)-N-benzyl-3,4-dihydroxy-5-methyl-D-prolinamide, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wright, D.W, Davies, G.J. | | Deposit date: | 2013-03-12 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Exploring a Multivalent Approach to alpha-L-Fucosidase Inhibition
EUR.J.ORG.CHEM., 2013, 2013
|
|
4MAH
 
 | | Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Zn | | Descriptor: | 1,2-ETHANEDIOL, AA11 Lytic Polysaccharide Monooxygenase, CHLORIDE ION, ... | | Authors: | Hemsworth, G.R, Henrissat, B, Walton, P.H, Davies, G.J. | | Deposit date: | 2013-08-16 | | Release date: | 2013-12-18 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and characterization of a new family of lytic polysaccharide monooxygenases.
Nat.Chem.Biol., 10, 2014
|
|
4MAI
 
 | | Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Cu(I) | | Descriptor: | 1,2-ETHANEDIOL, AA11 Lytic Polysaccharide Monooxygenase, CHLORIDE ION, ... | | Authors: | Hemsworth, G.R, Henrissat, B, Walton, P.H, Davies, G.J. | | Deposit date: | 2013-08-16 | | Release date: | 2013-12-18 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery and characterization of a new family of lytic polysaccharide monooxygenases.
Nat.Chem.Biol., 10, 2014
|
|
4OPB
 
 | | AA13 Lytic polysaccharide monooxygenase from Aspergillus oryzae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COPPER (II) ION, ... | | Authors: | Lo Leggio, L, Frandsen, K.H, Davies, G.J, Dupree, P, Walton, P, Henrissat, B, Stringer, M, Tovborg, M, De Maria, L, Johansen, K.S. | | Deposit date: | 2014-02-05 | | Release date: | 2015-01-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and boosting activity of a starch-degrading lytic polysaccharide monooxygenase.
Nat Commun, 6, 2015
|
|
7OU6
 
 | | Human O-GlcNAc hydrolase in complex with DNJNAc-thiazolidines | | Descriptor: | Protein O-GlcNAcase, ~{N}-[(3~{Z},6~{S},7~{R},8~{R},8~{a}~{S})-7,8-bis(oxidanyl)-3-(phenylmethyl)imino-1,5,6,7,8,8~{a}-hexahydro-[1,3]thiazolo[3,4-a]pyridin-6-yl]ethanamide | | Authors: | Males, A, Davies, G.J, Gonzalez-Cuesta, M, Mellet, C.O, Fernandez, J.M.G, Sidhu, P, Ashmus, R, Busmann, J, Vocadlo, D.J, Foster, L. | | Deposit date: | 2021-06-11 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Bicyclic Picomolar OGA Inhibitors Enable Chemoproteomic Mapping of Its Endogenous Post-translational Modifications
J.Am.Chem.Soc., 144, 2022
|
|
