9BUX
 
 | | Structure of GGCX-BGP-menaquinone-4 epoxide complex | | Descriptor: | (1aR,7aS)-1a-methyl-7a-[(2E,6E,10E)-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraen-1-yl]-1a,7a-dihydronaphtho[2,3-b]oxirene-2,7-dione, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, R, Qi, X. | | Deposit date: | 2024-05-17 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structure and mechanism of vitamin-K-dependent gamma-glutamyl carboxylase.
Nature, 639, 2025
|
|
9BUR
 
 | | Structure of GGCX-BGP complex | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, R, Qi, X. | | Deposit date: | 2024-05-17 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure and mechanism of vitamin-K-dependent gamma-glutamyl carboxylase.
Nature, 639, 2025
|
|
7YKJ
 
 | | Omicron RBDs bound with P3E6 Fab (one up and one down) | | Descriptor: | P3E6 heavy chain, P3E6 light chain, Spike glycoprotein | | Authors: | Tang, B, Dang, S. | | Deposit date: | 2022-07-22 | | Release date: | 2022-12-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into broadly neutralizing antibodies elicited by hybrid immunity against SARS-CoV-2.
Emerg Microbes Infect, 12, 2023
|
|
8H5U
 
 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain in complex with neutralizing nanobody Nb-021 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody Nb-021, ... | | Authors: | Yang, J, Lin, S, Lu, G.W. | | Deposit date: | 2022-10-13 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Development of a bispecific nanobody conjugate broadly neutralizes diverse SARS-CoV-2 variants and structural basis for its broad neutralization.
Plos Pathog., 19, 2023
|
|
8H5T
 
 | |
8K19
 
 | |
7YHH
 
 | |
7YHG
 
 | |
7YHF
 
 | |
7YHI
 
 | |
8W8G
 
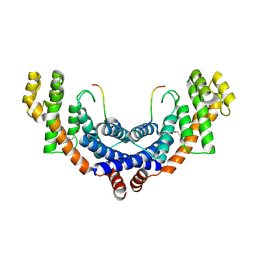 | | Crystal structure of human TRF1 with PinX1 | | Descriptor: | HEXANE-1,6-DIOL, PIN2/TERF1-interacting telomerase inhibitor 1, Telomeric repeat-binding factor 1 | | Authors: | Lei, M, Wu, J. | | Deposit date: | 2023-09-02 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | PinX1 suppresses cancer progression by inhibiting telomerase activity in cervical squamous cell carcinoma and endocervical adenocarcinoma.
Genes Dis, 12, 2025
|
|
7N64
 
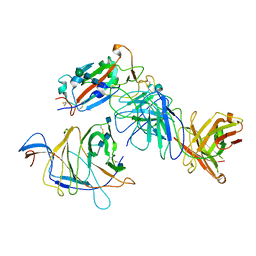 | | SARS-CoV-2 Spike (2P) in complex with G32R7 Fab (RBD and NTD local reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, G32R7 Fab heavy chain, ... | | Authors: | Windsor, I.W, Jenni, S, Tong, P, Gautam, A.K, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-06-07 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Memory B cell repertoire for recognition of evolving SARS-CoV-2 spike.
Biorxiv, 2021
|
|
7N62
 
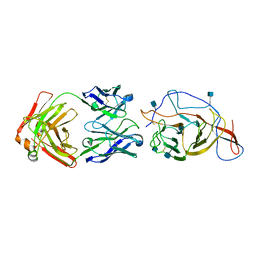 | | SARS-CoV-2 Spike (2P) in complex with C12C9 Fab (NTD local reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C12C9 Fab heavy chain, C12C9 Fab light chain, ... | | Authors: | Windsor, I.W, Jenni, S, Bajic, G, Tong, P, Gautam, A.K, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-06-07 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Memory B cell repertoire for recognition of evolving SARS-CoV-2 spike.
Biorxiv, 2021
|
|
7XH8
 
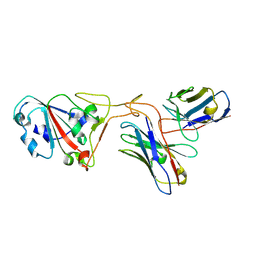 | | The structure of ZCB11 Fab against SARS-CoV-2 Omicron Spike | | Descriptor: | Spike glycoprotein, The heavy chain of ZCB11 antibody, The light chain of ZCB11 antibody | | Authors: | Hang, L, Dang, S. | | Deposit date: | 2022-04-07 | | Release date: | 2022-06-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | A broadly neutralizing antibody protects Syrian hamsters against SARS-CoV-2 Omicron challenge.
Nat Commun, 13, 2022
|
|
7Y1F
 
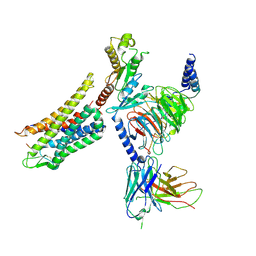 | | Cryo-EM structure of human k-opioid receptor-Gi complex | | Descriptor: | Dynorphin, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Chen, B.O, Xu, F.E. | | Deposit date: | 2022-06-08 | | Release date: | 2023-05-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of human kappa-opioid receptor-Gi complex bound to an endogenous agonist dynorphin A.
Protein Cell, 14, 2023
|
|
7W1S
 
 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain in complex with neutralizing nanobody Nb-007 | | Descriptor: | Nanobody Nb-007, Spike protein S1 | | Authors: | Yang, J, Lin, S, Sun, H.L, Lu, G.W. | | Deposit date: | 2021-11-20 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | A Potent Neutralizing Nanobody Targeting the Spike Receptor-Binding Domain of SARS-CoV-2 and the Structural Basis of Its Intimate Binding.
Front Immunol, 13, 2022
|
|
7DK0
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW05 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MW05 heavy chain, MW05 light chain, ... | | Authors: | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-11-22 | | Release date: | 2021-06-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.199 Å) | | Cite: | Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires Fc gamma RIIB and virus-antibody complex with bivalent interaction.
Commun Biol, 5, 2022
|
|
7DJZ
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW01 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, MW01 heavy chain, ... | | Authors: | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-11-22 | | Release date: | 2021-06-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires Fc gamma RIIB and virus-antibody complex with bivalent interaction.
Commun Biol, 5, 2022
|
|
9ARJ
 
 | |
9ARK
 
 | |
9ARL
 
 | |
3J3Y
 
 | |
3J4F
 
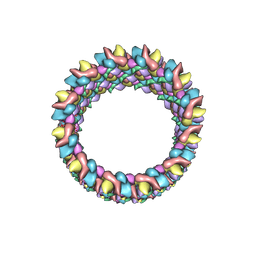 | | Structure of HIV-1 capsid protein by cryo-EM | | Descriptor: | capsid protein | | Authors: | Zhao, G, Perilla, J.R, Meng, X, Schulten, K, Zhang, P. | | Deposit date: | 2013-07-11 | | Release date: | 2013-07-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics.
Nature, 497, 2013
|
|
3J3Q
 
 | |
8XFW
 
 | |
