6TRM
 
 | | Solution structure of the antifungal protein PAFC | | Descriptor: | Pc21g12970 protein | | Authors: | Czajlik, A, Holzknecht, J, Marx, F, Batta, G. | | Deposit date: | 2019-12-19 | | Release date: | 2020-10-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Dynamics, and New Antifungal Aspects of the Cysteine-Rich Miniprotein PAFC.
Int J Mol Sci, 22, 2021
|
|
2YCM
 
 | | Inhibitors of herbicidal target IspD | | Descriptor: | 2-C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLYLTRANSFERASE, CHLOROPLASTIC, 6-BENZYL-5-CHLORO-7-HYDROXYPYRAZOLO[1,5-A]PYRIMIDINE-3-CARBOXYLIC ACID, ... | | Authors: | Hoeffken, H.W. | | Deposit date: | 2011-03-16 | | Release date: | 2011-08-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibitors of the Herbicidal Target Ispd: Allosteric Site Binding.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
2YC5
 
 | | Inhibitors of the herbicidal target IspD | | Descriptor: | 2-C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLYLTRANSFERASE, CHLOROPLASTIC, 5-chloro-7-hydroxy-6-(phenylmethyl)pyrazolo[1,5-a]pyrimidine-3-carbonitrile, ... | | Authors: | Hoeffken, H.W. | | Deposit date: | 2011-03-11 | | Release date: | 2011-08-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibitors of the Herbicidal Target Ispd: Allosteric Site Binding.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
4ZX2
 
 | | Co-crystal structures of PP5 in complex with 5-methyl-7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid | | Descriptor: | (1S,2R,3S,4R,5S)-5-methyl-7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | Chattopadhyay, D, Swingle, M.R, Salter, E.A, Wierzbicki, A, Honkanen, R.E. | | Deposit date: | 2015-05-19 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Crystal structures and mutagenesis of PPP-family ser/thr protein phosphatases elucidate the selectivity of cantharidin and novel norcantharidin-based inhibitors of PP5C.
Biochem. Pharmacol., 109, 2016
|
|
4ZVZ
 
 | | Co-crystal structures of PP5 in complex with 5-methyl-7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid | | Descriptor: | (1R,2S,3R,4S,5S)-5-(propoxymethyl)-7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid, DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, ... | | Authors: | Chattopadhyay, D, Swingle, M.R, Salter, E.A, Wierzbicki, A, Honkanen, R.E. | | Deposit date: | 2015-05-18 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and mutagenesis of PPP-family ser/thr protein phosphatases elucidate the selectivity of cantharidin and novel norcantharidin-based inhibitors of PP5C.
Biochem. Pharmacol., 109, 2016
|
|
8CN1
 
 | | hDLG1-PDZ1 in complex with a TAX1 peptide from HTLV-1 | | Descriptor: | Disks large homolog 1, GLU-THR-GLU-VAL, SULFATE ION | | Authors: | Maseko, S, Sogues, A, Volkov, A, Remaut, H, Twizere, J.C. | | Deposit date: | 2023-02-21 | | Release date: | 2023-08-02 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Identification of small molecule antivirals against HTLV-1 by targeting the hDLG1-Tax-1 protein-protein interaction.
Antiviral Res., 217, 2023
|
|
8CN3
 
 | | hDLG1-PDZ2 in complex with a TAX1 peptide from HTLV-1 | | Descriptor: | Disks large homolog 1, GLU-THR-GLU-VAL | | Authors: | Maseko, S, Sogues, A, Volkov, A, Remaut, H, Twizere, J.C. | | Deposit date: | 2023-02-21 | | Release date: | 2023-08-02 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Identification of small molecule antivirals against HTLV-1 by targeting the hDLG1-Tax-1 protein-protein interaction.
Antiviral Res., 217, 2023
|
|
7R8J
 
 | |
7R8F
 
 | |
7R8K
 
 | |
7R8G
 
 | |
7R8H
 
 | |
7AK0
 
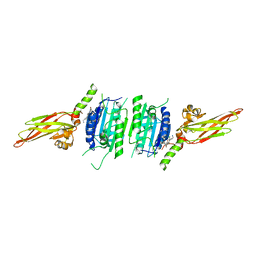 | | Human MALT1(329-729) in complex with a chromane urea containing inhibitor | | Descriptor: | 1-[4-[4-(aminomethyl)pyrazol-1-yl]-3-chloranyl-phenyl]-3-[(3~{R})-6-bromanyl-3,4-dihydro-2~{H}-chromen-3-yl]urea, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Renatus, M. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.316 Å) | | Cite: | Discovery of Potent, Highly Selective, and In Vivo Efficacious, Allosteric MALT1 Inhibitors by Iterative Scaffold Morphing.
J.Med.Chem., 63, 2020
|
|
7AK1
 
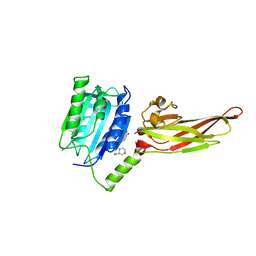 | | Human MALT1(329-729) in complex with a chromane urea containing inhibitor | | Descriptor: | 1-(3-chloranyl-4-methoxy-phenyl)-3-[7-[(3~{S})-3-(methoxymethyl)morpholin-4-yl]-2-methyl-pyrazolo[1,5-a]pyrimidin-6-yl]urea, MAGNESIUM ION, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Renatus, M. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | Discovery of Potent, Highly Selective, and In Vivo Efficacious, Allosteric MALT1 Inhibitors by Iterative Scaffold Morphing.
J.Med.Chem., 63, 2020
|
|
6Q9L
 
 | |
6Q9U
 
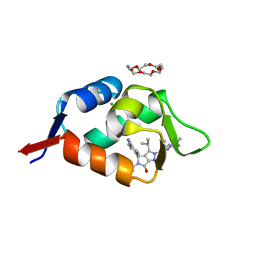 | | HDMX (14-111; C17S) COMPLEXED WITH COMPOUND 12 AT 2.4A; Structural states of Hdm2 and HdmX: X-ray elucidation of adaptations and binding interactions for different chemical compound classes | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 4-[(4~{S})-5-(5-chloranyl-2-oxidanylidene-1~{H}-pyridin-3-yl)-2-[2-(dimethylamino)-4-methoxy-pyrimidin-5-yl]-6-oxidanylidene-3-propan-2-yl-4~{H}-pyrrolo[3,4-c]pyrazol-4-yl]benzenecarbonitrile, CHLORIDE ION, ... | | Authors: | Kallen, J. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural States of Hdm2 and HdmX: X-ray Elucidation of Adaptations and Binding Interactions for Different Chemical Compound Classes.
Chemmedchem, 14, 2019
|
|
6Q9O
 
 | |
6Q9Q
 
 | | HDMX (14-111; C17S) COMPLEXED WITH COMPOUND 13 AT 2.1A; Structural states of Hdm2 and HdmX: X-ray elucidation of adaptations and binding interactions for different chemical compound classes | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 6-chloranyl-3-[3-[(1~{S})-1-(4-chlorophenyl)ethyl]-5-phenyl-imidazol-4-yl]-~{N}-[2-(4-cyclohexylpiperazin-1-yl)ethyl]-1~{H}-indole-2-carboxamide, GLYCEROL, ... | | Authors: | Kallen, J. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural States of Hdm2 and HdmX: X-ray Elucidation of Adaptations and Binding Interactions for Different Chemical Compound Classes.
Chemmedchem, 14, 2019
|
|
6Q9W
 
 | | X-ray structure of compound 15 bound to HdmX: Structural states of Hdm2 and HdmX: X-ray elucidation of adaptations and binding interactions for different chemical compound classes | | Descriptor: | (4~{S})-4-(4-chlorophenyl)-5-[(1~{S})-1-(3-chlorophenyl)ethyl]-2-(2,4-dimethoxypyrimidin-5-yl)-3-propan-2-yl-4~{H}-pyrrolo[3,4-d]imidazol-6-one, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Protein Mdm4, ... | | Authors: | Kallen, J. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural States of Hdm2 and HdmX: X-ray Elucidation of Adaptations and Binding Interactions for Different Chemical Compound Classes.
Chemmedchem, 14, 2019
|
|
6Q9H
 
 | |
6Q9Y
 
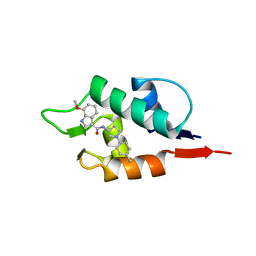 | |
6Q9S
 
 | | HDMX (14-111; C17S) COMPLEXED WITH COMPOUND 14 AT 2.4A: Structural states of Hdm2 and HdmX: X-ray elucidation of adaptations and binding interactions for different chemical compound classes | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 3-[[6-chloranyl-3-[3-[(1~{S})-1-(2,4-dichlorophenyl)ethyl]-5-phenyl-imidazol-4-yl]-1~{H}-indol-2-yl]carbonylamino]-4-[4-(2-oxidanylidene-1,3-oxazinan-3-yl)piperidin-1-yl]benzoic acid, Protein Mdm4, ... | | Authors: | Kallen, J. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural States of Hdm2 and HdmX: X-ray Elucidation of Adaptations and Binding Interactions for Different Chemical Compound Classes.
Chemmedchem, 14, 2019
|
|
6Q96
 
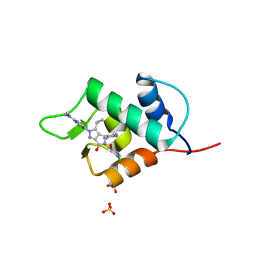 | |
5K59
 
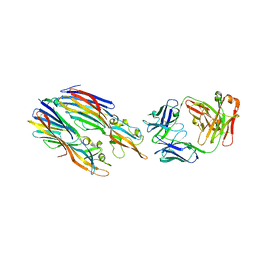 | | Crystal structure of LukGH from Staphylococcus aureus in complex with a neutralising antibody | | Descriptor: | CHLORIDE ION, Fab heavy chain, Fab light chain, ... | | Authors: | Welin, M, Logan, D.T, Badarau, A, Mirkina, I, Zauner, G, Dolezilkova, I, Nagy, E. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-10 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Context matters: The importance of dimerization-induced conformation of the LukGH leukocidin of Staphylococcus aureus for the generation of neutralizing antibodies.
Mabs, 8, 2016
|
|
6YN8
 
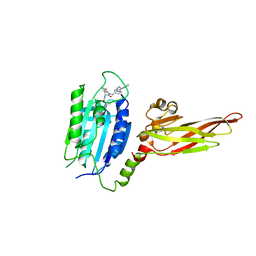 | | Human MALT1(334-719) in complex with a tetrazole containing compound | | Descriptor: | 3-azanyl-3-methyl-~{N}-[(3~{R})-4-oxidanylidene-5-[[4-[2-(1~{H}-1,2,3,4-tetrazol-5-yl)phenyl]phenyl]methyl]-2,3-dihydro-1,5-benzoxazepin-3-yl]butanamide, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Renatus, M. | | Deposit date: | 2020-04-11 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Stabilizing Inactive Conformations of MALT1 as an Effective Approach to Inhibit Its Protease Activity
Advanced Therapeutics, 3, 2020
|
|
