9EQF
 
 | | Crystal structure of the L-arginine hydroxylase VioC MeHis316, bound to Fe(II), L-arginine, and succinate | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, Alpha-ketoglutarate-dependent L-arginine hydroxylase, ... | | Authors: | Hardy, F.J. | | Deposit date: | 2024-03-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing Ferryl Reactivity in a Nonheme Iron Oxygenase Using an Expanded Genetic Code.
Acs Catalysis, 14, 2024
|
|
9F18
 
 | |
9FTW
 
 | |
9C0O
 
 | |
9ARD
 
 | |
7PQO
 
 | | Catalytic fragment of MASP-1 in complex with P1 site mutant ecotin | | Descriptor: | Ecotin, GLYCEROL, Mannan-binding lectin serine protease 1, ... | | Authors: | Harmat, V, Fodor, K, Heja, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Synergy of protease-binding sites within the ecotin homodimer is crucial for inhibition of MASP enzymes and for blocking lectin pathway activation.
J.Biol.Chem., 298, 2022
|
|
1F5J
 
 | | CRYSTAL STRUCTURE OF XYNB, A HIGHLY THERMOSTABLE BETA-1,4-XYLANASE FROM DICTYOGLOMUS THERMOPHILUM RT46B.1, AT 1.8 A RESOLUTION | | Descriptor: | BETA-1,4-XYLANASE, SULFATE ION | | Authors: | McCarthy, A.A, Baker, E.N. | | Deposit date: | 2000-07-26 | | Release date: | 2000-11-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of XynB, a highly thermostable beta-1,4-xylanase from Dictyoglomus thermophilum Rt46B.1, at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1FL1
 
 | | KSHV PROTEASE | | Descriptor: | POTASSIUM ION, PROTEASE | | Authors: | Reiling, K.K, Pray, T.R, Craik, C.S, Stroud, R.M. | | Deposit date: | 2000-08-11 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional consequences of the Kaposi's sarcoma-associated herpesvirus protease structure: regulation of activity and dimerization by conserved structural elements.
Biochemistry, 39, 2000
|
|
6EYC
 
 | | Re-refinement of the MCM2-7 double hexamer using ISOLDE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, ... | | Authors: | Croll, T.I. | | Deposit date: | 2017-11-11 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
8OXU
 
 | | Crystal Structure of the Hsp90-LA1011 Complex | | Descriptor: | ATP-dependent molecular chaperone HSP82, dimethyl 2,6-bis[2-(dimethylamino)ethyl]-1-methyl-4-[4-(trifluoromethyl)phenyl]-4~{H}-pyridine-3,5-dicarboxylate | | Authors: | Roe, S.M, Prodromou, C. | | Deposit date: | 2023-05-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | The Crystal Structure of the Hsp90-LA1011 Complex and the Mechanism by Which LA1011 May Improve the Prognosis of Alzheimer's Disease.
Biomolecules, 13, 2023
|
|
6FOY
 
 | |
6FP1
 
 | | The crystal structure of P.fluorescens Kynurenine 3-monooxygenase (KMO) in complex with competitive inhibitor No. 1 | | Descriptor: | 2-(6-chloranyl-5,7-dimethyl-3-oxidanylidene-1,4-benzoxazin-4-yl)ethanoic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Levy, C.W, Leys, D. | | Deposit date: | 2018-02-08 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A brain-permeable inhibitor of the neurodegenerative disease target kynurenine 3-monooxygenase prevents accumulation of neurotoxic metabolites.
Commun Biol, 2, 2019
|
|
6FP0
 
 | |
6FOX
 
 | |
7C7J
 
 | |
7C7I
 
 | |
1KP8
 
 | |
1LW1
 
 | |
1N51
 
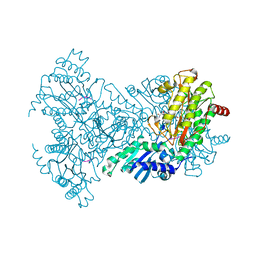 | | Aminopeptidase P in complex with the inhibitor apstatin | | Descriptor: | MANGANESE (II) ION, Xaa-Pro aminopeptidase, apstatin | | Authors: | Graham, S.C, Maher, M.J, Lee, M.H, Simmons, W.H, Freeman, H.C, Guss, J.M. | | Deposit date: | 2002-11-03 | | Release date: | 2003-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Escherichia coli aminopeptidase P in complex with the inhibitor apstatin.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1MNF
 
 | |
1OWM
 
 | | DATA1:DNA photolyase / received X-rays dose 1.2 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OWN
 
 | | DATA3:DNA photolyase / received X-rays dose 4.8 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1ZXJ
 
 | | Crystal structure of the hypthetical Mycoplasma protein, MPN555 | | Descriptor: | Hypothetical protein MG377 homolog | | Authors: | Schulze-Gahmen, U, Aono, S, Shengfeng, C, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2005-06-08 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the hypothetical Mycoplasma protein MPN555 suggests a chaperone function.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
7PQN
 
 | | Catalytic fragment of MASP-2 in complex with ecotin | | Descriptor: | Ecotin, GLYCEROL, Mannan-binding lectin serine protease 2 A chain, ... | | Authors: | Harmat, V, Fodor, K, Heja, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.400015 Å) | | Cite: | Synergy of protease-binding sites within the ecotin homodimer is crucial for inhibition of MASP enzymes and for blocking lectin pathway activation.
J.Biol.Chem., 298, 2022
|
|
6PW8
 
 | |
