6VSO
 
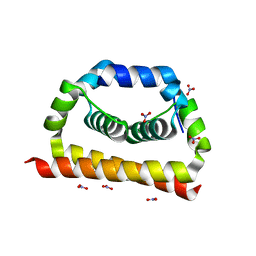 | | DengueV-2 Capsid Structure | | Descriptor: | Capsid premembrane protein, GLYCEROL, NITRATE ION | | Authors: | White, M, Xia, H, Shi, P. | | Deposit date: | 2020-02-11 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | A cocrystal structure of dengue capsid protein in complex of inhibitor.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8TMA
 
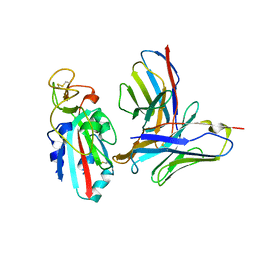 | | Antibody N3-1 bound to RBD in the up conformation | | Descriptor: | N3-1 Fab heavy chain, N3-1 Fab light chain, Spike glycoprotein | | Authors: | Hsieh, C.-L, McLellan, J.S. | | Deposit date: | 2023-07-29 | | Release date: | 2024-01-17 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | SARS-COV-2 Omicron variants conformationally escape a rare quaternary antibody binding mode.
Commun Biol, 6, 2023
|
|
8TM1
 
 | |
3G9N
 
 | |
3I4B
 
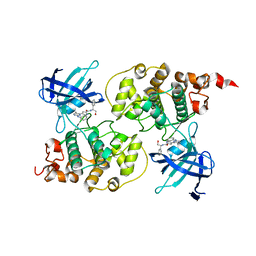 | | Crystal structure of GSK3b in complex with a pyrimidylpyrrole inhibitor | | Descriptor: | Glycogen synthase kinase-3 beta, N-[(1S)-2-hydroxy-1-phenylethyl]-4-[5-methyl-2-(phenylamino)pyrimidin-4-yl]-1H-pyrrole-2-carboxamide | | Authors: | Ter Haar, E. | | Deposit date: | 2009-07-01 | | Release date: | 2010-01-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided design of potent and selective pyrimidylpyrrole inhibitors of extracellular signal-regulated kinase (ERK) using conformational control.
J.Med.Chem., 52, 2009
|
|
3G9L
 
 | |
4MBS
 
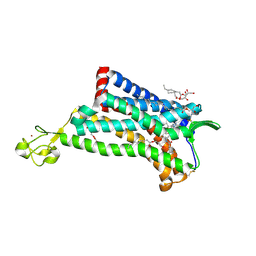 | | Crystal Structure of the CCR5 Chemokine Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4,4-difluoro-N-[(1S)-3-{(3-exo)-3-[3-methyl-5-(propan-2-yl)-4H-1,2,4-triazol-4-yl]-8-azabicyclo[3.2.1]oct-8-yl}-1-phenylpropyl]cyclohexanecarboxamide, Chimera protein of C-C chemokine receptor type 5 and Rubredoxin, ... | | Authors: | Tan, Q, Zhu, Y, Han, G.W, Li, J, Fenalti, G, Liu, H, Cherezov, V, Stevens, R.C, GPCR Network (GPCR), Zhao, Q, Wu, B. | | Deposit date: | 2013-08-19 | | Release date: | 2013-09-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex.
Science, 341, 2013
|
|
9GGC
 
 | |
9GGB
 
 | |
9GGF
 
 | |
9GGE
 
 | |
9GGD
 
 | |
9KHQ
 
 | | Crystal structure of N-acyl homoserine lactonase AhlX | | Descriptor: | MAGNESIUM ION, N-acylhomoserine lactonase, NICKEL (II) ION | | Authors: | Chen, Y, Chu, X.H. | | Deposit date: | 2024-11-11 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An AHL-lactonase mutant featuring a unique "tri-His" motif exhibits enhanced activity, stability and effectively controls plant soft rot.
Int.J.Biol.Macromol., 308, 2025
|
|
9KHO
 
 | |
8J19
 
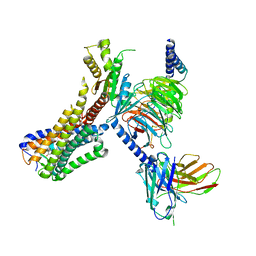 | | Cryo-EM structure of the LY237-bound GPR84 receptor-Gi complex | | Descriptor: | 6-nonylpyridine-2,4-diol, Antibody fragment ScFv16, G-protein coupled receptor 84, ... | | Authors: | Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into ligand recognition and activation of the medium-chain fatty acid-sensing receptor GPR84.
Nat Commun, 14, 2023
|
|
8J1A
 
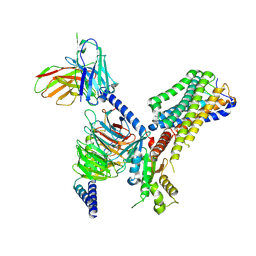 | | Cryo-EM structure of the GPR84 receptor-Gi complex with no ligand modeled | | Descriptor: | Antibody fragment ScFv16, G-protein coupled receptor 84, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural insights into ligand recognition and activation of the medium-chain fatty acid-sensing receptor GPR84.
Nat Commun, 14, 2023
|
|
8J18
 
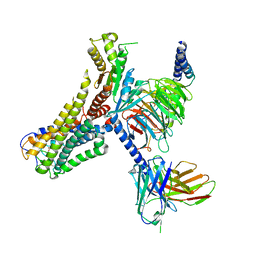 | | Cryo-EM structure of the 3-OH-C12-bound GPR84 receptor-Gi complex | | Descriptor: | (3R)-3-HYDROXYDODECANOIC ACID, Antibody fragment ScFv16, G-protein coupled receptor 84, ... | | Authors: | Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural insights into ligand recognition and activation of the medium-chain fatty acid-sensing receptor GPR84.
Nat Commun, 14, 2023
|
|
4JBR
 
 | |
8WD8
 
 | | Cryo-EM structure of TtdAgo-guide DNA-target DNA complex | | Descriptor: | Argonaute family protein, Guide DNA, MAGNESIUM ION, ... | | Authors: | Zhuang, L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-01-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanism for target recognition, dimerization, and activation of Pyrococcus furiosus Argonaute.
Mol.Cell, 84, 2024
|
|
8X17
 
 | | Cryo-EM structure of adenosine receptor A3AR bound to CF102 | | Descriptor: | Adenosine receptor A3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cai, H, Xu, Y, Xu, H.E. | | Deposit date: | 2023-11-06 | | Release date: | 2024-04-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Cryo-EM structures of adenosine receptor A 3 AR bound to selective agonists.
Nat Commun, 15, 2024
|
|
8X16
 
 | | Cryo-EM structure of adenosine receptor A3AR bound to CF101 | | Descriptor: | Adenosine receptor A3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cai, H, Xu, Y, Xu, H.E. | | Deposit date: | 2023-11-06 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structures of adenosine receptor A 3 AR bound to selective agonists.
Nat Commun, 15, 2024
|
|
5I3P
 
 | | DENGUE SEROTYPE 3 RNA-DEPENDENT RNA POLYMERASE BOUND TO COMPOUND 27 | | Descriptor: | 5-[5-(3-hydroxyprop-1-yn-1-yl)thiophen-2-yl]-2,4-dimethoxy-N-[(3-methoxyphenyl)sulfonyl]benzamide, Genome polyprotein, ZINC ION | | Authors: | Noble, C.G. | | Deposit date: | 2016-02-10 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Potent Allosteric Dengue Virus NS5 Polymerase Inhibitors: Mechanism of Action and Resistance Profiling
Plos Pathog., 12, 2016
|
|
5I3Q
 
 | | DENGUE SEROTYPE 3 RNA-DEPENDENT RNA POLYMERASE BOUND TO COMPOUND 29 | | Descriptor: | 5-[5-(3-hydroxyprop-1-yn-1-yl)thiophen-2-yl]-4-methoxy-2-methyl-N-[(quinolin-8-yl)sulfonyl]benzamide, Genome polyprotein, ZINC ION | | Authors: | Noble, C.G. | | Deposit date: | 2016-02-10 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Potent Allosteric Dengue Virus NS5 Polymerase Inhibitors: Mechanism of Action and Resistance Profiling
Plos Pathog., 12, 2016
|
|
5JJR
 
 | | Dengue 3 NS5 protein with compound 29 | | Descriptor: | 1,2-ETHANEDIOL, 5-[5-(3-hydroxyprop-1-yn-1-yl)thiophen-2-yl]-4-methoxy-2-methyl-N-[(quinolin-8-yl)sulfonyl]benzamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lescar, J, El Sahili, A. | | Deposit date: | 2016-04-25 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Potent Allosteric Dengue Virus NS5 Polymerase Inhibitors: Mechanism of Action and Resistance Profiling
Plos Pathog., 12, 2016
|
|
6A7U
 
 | | Crystal structure of histone H2A.Bbd-H2B dimer | | Descriptor: | Histone H2B type 2-E,Histone H2A-Bbd type 2/3 | | Authors: | Dai, L, Zhou, Z. | | Deposit date: | 2018-07-04 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the histone heterodimer containing histone variant H2A.Bbd.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
