8ROX
 
 | | Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 12 | | Descriptor: | 5-[[3,4-bis(chloranyl)-1~{H}-indol-7-yl]sulfamoyl]-~{N},~{N},3-trimethyl-furan-2-carboxamide;ethane, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Shilliday, F, Lucas, S.C.C, Richter, M, Michaelides, I.N, Fusani, L. | | Deposit date: | 2024-01-12 | | Release date: | 2024-04-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Optimization of Potent Ligands for the E3 Ligase DCAF15 and Evaluation of Their Use in Heterobifunctional Degraders.
J.Med.Chem., 67, 2024
|
|
7RW2
 
 | |
5DH3
 
 | | Crystal structure of MST2 in complex with XMU-MP-1 | | Descriptor: | 4-[(5,10-dimethyl-6-oxo-6,10-dihydro-5H-pyrimido[5,4-b]thieno[3,2-e][1,4]diazepin-2-yl)amino]benzenesulfonamide, CHLORIDE ION, SULFATE ION, ... | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2015-08-29 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.468 Å) | | Cite: | Pharmacological targeting of kinases MST1 and MST2 augments tissue repair and regeneration
Sci Transl Med, 8, 2016
|
|
8HLE
 
 | | Structure of DddY-DMSOP complex | | Descriptor: | 3-[dimethyl(oxidanyl)-$l^{4}-sulfanyl]propanoic acid, DMSP lyase DddY, ZINC ION | | Authors: | Peng, M, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2022-11-30 | | Release date: | 2023-10-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | DMSOP-cleaving enzymes are diverse and widely distributed in marine microorganisms.
Nat Microbiol, 8, 2023
|
|
8HLF
 
 | | Crystal structure of DddK-DMSOP complex | | Descriptor: | 3-[dimethyl(oxidanyl)-$l^{4}-sulfanyl]propanoic acid, MANGANESE (II) ION, Novel protein with potential Cupin domain | | Authors: | Peng, M, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2022-11-30 | | Release date: | 2023-10-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | DMSOP-cleaving enzymes are diverse and widely distributed in marine microorganisms.
Nat Microbiol, 8, 2023
|
|
4OY2
 
 | | Crystal structure of TAF1-TAF7, a TFIID subcomplex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 7, ... | | Authors: | Bhattacharya, S, Lou, X, Rajashankar, K, Jacobson, R.H, Webb, P. | | Deposit date: | 2014-02-10 | | Release date: | 2014-06-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional insight into TAF1-TAF7, a subcomplex of transcription factor II D.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7F3B
 
 | | cocrystallization of Escherichia coli dihydrofolate reductase (DHFR) and its pyrrolo[3,2-f]quinazoline inhibitor. | | Descriptor: | 7-[(2-fluorophenyl)methyl]pyrrolo[3,2-f]quinazoline-1,3-diamine, Dihydrofolate reductase, GLYCEROL | | Authors: | Wang, H, You, X.F, Yang, X.Y, Li, Y, Hong, W. | | Deposit date: | 2021-06-16 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The discovery of 1, 3-diamino-7H-pyrrol[3, 2-f]quinazoline compounds as potent antimicrobial antifolates.
Eur.J.Med.Chem., 228, 2022
|
|
6F57
 
 | |
5XLO
 
 | |
6KV5
 
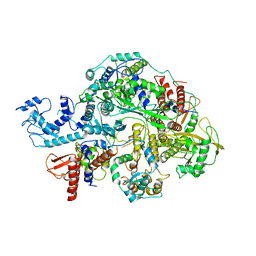 | | Structure of influenza D virus apo polymerase | | Descriptor: | Polymerase 3, Polymerase PB2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6MD9
 
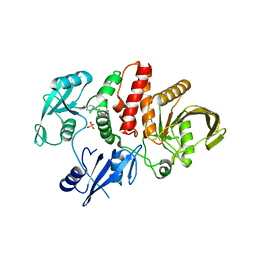 | |
6MDA
 
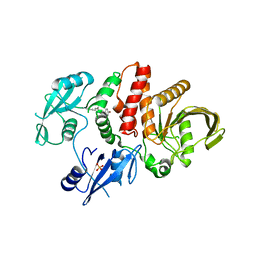 | |
6M7V
 
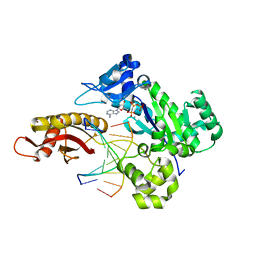 | | Human DNA polymerase eta extension complex with cdA at the -1 position | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(P*(02I)P*CP*TP*CP*AP*CP*AP*CP*T)-3'), DNA (5'-D(P*GP*TP*GP*TP*GP*AP*GP*T)-3'), ... | | Authors: | Weng, P, Gao, Y, Yang, W. | | Deposit date: | 2018-08-21 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.062 Å) | | Cite: | Bypassing a 8,5'-cyclo-2'-deoxyadenosine lesion by human DNA polymerase eta at atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6M7O
 
 | | Human DNA polymerase eta ternary complex with Mn2+ and dTMPNPP oppositing cdA | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA (5'-D(*AP*TP*(02I)P*CP*TP*CP*AP*CP*AP*CP*T)-3'), DNA (5'-D(P*AP*GP*TP*GP*TP*GP*AP*G*(1FZ))-3'), ... | | Authors: | Weng, P, Gao, Y, Yang, W. | | Deposit date: | 2018-08-20 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bypassing a 8,5'-cyclo-2'-deoxyadenosine lesion by human DNA polymerase eta at atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7ESI
 
 | | Crystal structure of the collagenase unit of a Vibrio collagenase from Vibrio harveyi VHJR7 at 1. 8 angstrom resolution. | | Descriptor: | CALCIUM ION, Collagenase unit (CU), Peptide P1, ... | | Authors: | Cao, H.Y, Wang, Y, Peng, M, Zhang, Y.Z. | | Deposit date: | 2021-05-11 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Vibrio collagenase VhaC provides insight into the mechanism of bacterial collagenolysis.
Nat Commun, 13, 2022
|
|
9MRE
 
 | |
7FIL
 
 | |
7FIK
 
 | | The cryo-EM structure of the CR subunit from X. laevis NPC | | Descriptor: | MGC154553 protein, MGC83295 protein, MGC83926 protein, ... | | Authors: | Shi, Y, Huang, G, Zhan, X. | | Deposit date: | 2021-07-31 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex.
Science, 376, 2022
|
|
5HRR
 
 | | HIV Integrase Catalytic Domain containing F185K + A124N + T125S mutations complexed with GSK0002 | | Descriptor: | (2S)-tert-butoxy[1-(3,4-difluorobenzyl)-6-methyl-4-(5-methyl-3,4-dihydro-2H-chromen-6-yl)-1H-pyrrolo[2,3-b]pyridin-5-yl]acetic acid, 1,2-ETHANEDIOL, CACODYLATE ION, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2016-01-24 | | Release date: | 2016-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Basis for Inhibitor-Induced Aggregation of HIV Integrase.
PLoS Biol., 14, 2016
|
|
5HRP
 
 | | HIV Integrase Catalytic Domain containing F185K + A124T mutations complexed with GSK0002 | | Descriptor: | (2S)-tert-butoxy[1-(3,4-difluorobenzyl)-6-methyl-4-(5-methyl-3,4-dihydro-2H-chromen-6-yl)-1H-pyrrolo[2,3-b]pyridin-5-yl]acetic acid, 1,2-ETHANEDIOL, CACODYLATE ION, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2016-01-24 | | Release date: | 2016-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Basis for Inhibitor-Induced Aggregation of HIV Integrase.
PLoS Biol., 14, 2016
|
|
5HRS
 
 | | HIV Integrase Catalytic Domain containing F185K + A124N + T125A mutations complexed with GSK0002 | | Descriptor: | (2S)-tert-butoxy[1-(3,4-difluorobenzyl)-6-methyl-4-(5-methyl-3,4-dihydro-2H-chromen-6-yl)-1H-pyrrolo[2,3-b]pyridin-5-yl]acetic acid, 1,2-ETHANEDIOL, CACODYLATE ION, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2016-01-24 | | Release date: | 2016-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Basis for Inhibitor-Induced Aggregation of HIV Integrase.
PLoS Biol., 14, 2016
|
|
5HRN
 
 | | HIV Integrase Catalytic Domain containing F185K mutation complexed with GSK0002 | | Descriptor: | (2S)-tert-butoxy[1-(3,4-difluorobenzyl)-6-methyl-4-(5-methyl-3,4-dihydro-2H-chromen-6-yl)-1H-pyrrolo[2,3-b]pyridin-5-yl]acetic acid, 1,2-ETHANEDIOL, CACODYLATE ION, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2016-01-23 | | Release date: | 2016-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Inhibitor-Induced Aggregation of HIV Integrase.
PLoS Biol., 14, 2016
|
|
3KCU
 
 | | Structure of formate channel | | Descriptor: | 2-(6-(2-CYCLOHEXYLETHOXY)-TETRAHYDRO-4,5-DIHYDROXY-2(HYDROXYMETHYL)-2H-PYRAN-3-YLOXY)-TETRAHYDRO-6(HYDROXYMETHYL)-2H-PY RAN-3,4,5-TRIOL, Probable formate transporter 1 | | Authors: | Wang, Y, Huang, Y, Wang, J, Yan, N, Shi, Y. | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | Structure of the formate transporter FocA reveals a pentameric aquaporin-like channel
Nature, 462, 2009
|
|
3KCV
 
 | | Structure of formate channel | | Descriptor: | Probable formate transporter 1 | | Authors: | Wang, Y, Huang, Y, Wang, J, Yan, N, Shi, Y. | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.198 Å) | | Cite: | Structure of the formate transporter FocA reveals a pentameric aquaporin-like channel
Nature, 462, 2009
|
|
3SFH
 
 | | Crystal Structure of Human HDAC8 Inhibitor Complex, an Amino Acid Derived Inhibitor | | Descriptor: | (2R)-2-amino-3-(2,4-dichlorophenyl)-1-(1,3-dihydro-2H-isoindol-2-yl)propan-1-one, ACETATE ION, Histone deacetylase 8, ... | | Authors: | Stams, T, Vash, B. | | Deposit date: | 2011-06-13 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human HDAC isoform selectivity achieved via exploitation of the acetate release channel with structurally unique small molecule inhibitors.
Bioorg.Med.Chem., 19, 2011
|
|
