2XMS
 
 | | Crystal structure of human NDRG2 protein provides insight into its role as a tumor suppressor | | Descriptor: | CHLORIDE ION, IMIDAZOLE, PROTEIN NDRG2 | | Authors: | Hwang, J, Kim, Y, Lee, H, Kim, M.H. | | Deposit date: | 2010-07-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Human Ndrg2 Protein Provides Insight Into its Role as a Tumor Suppressor.
J.Biol.Chem., 286, 2011
|
|
2XMR
 
 | | Crystal structure of human NDRG2 protein provides insight into its role as a tumor suppressor | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Hwang, J, Kim, Y, Lee, H, Kim, M.H. | | Deposit date: | 2010-07-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Ndrg2 Protein Provides Insight Into its Role as a Tumor Suppressor.
J.Biol.Chem., 286, 2011
|
|
8HEA
 
 | | Esterase2 (EaEst2) from Exiguobacterium antarcticum | | Descriptor: | Thermostable carboxylesterase Est30 | | Authors: | Hwang, J, Lee, J.H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and Biochemical Insights into Bis(2-hydroxyethyl) Terephthalate Degrading Carboxylesterase Isolated from Psychrotrophic Bacterium Exiguobacterium antarcticum.
Int J Mol Sci, 24, 2023
|
|
8HM5
 
 | |
5DQY
 
 | | A fully oxidized human thioredoxin | | Descriptor: | BENZOIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Hwang, J. | | Deposit date: | 2015-09-15 | | Release date: | 2015-12-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of fully oxidized human thioredoxin.
Biochem.Biophys.Res.Commun., 467, 2015
|
|
8ILJ
 
 | |
8JUO
 
 | |
8JUS
 
 | |
6LVO
 
 | |
7YVT
 
 | |
4IVN
 
 | | The Vibrio vulnificus NanR protein complexed with ManNAc-6P | | Descriptor: | 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-mannopyranose, Transcriptional regulator | | Authors: | Hwang, J, Kim, M.H. | | Deposit date: | 2013-01-23 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the regulation of sialic acid catabolism by the Vibrio vulnificus transcriptional repressor NanR
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4V24
 
 | | Sphingosine kinase 1 in complex with PF-543 | | Descriptor: | ACETATE ION, SPHINGOSINE KINASE 1, {(2R)-1-[4-({3-METHYL-5-[(PHENYLSULFONYL)METHYL]PHENOXY}METHYL)BENZYL]PYRROLIDIN-2-YL}METHANOL | | Authors: | Elkins, J.M, Wang, J, Sorrell, F, Tallant, C, Wang, D, Shrestha, L, Bountra, C, von Delft, F, Knapp, S, Edwards, A. | | Deposit date: | 2014-10-05 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Sphingosine Kinase 1 with Pf-543.
Acs Med.Chem.Lett., 5, 2014
|
|
5ED3
 
 | | crystal structure of human Hint1 complexing with AP5A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2015-10-20 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.309 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
6NXF
 
 | |
8IFF
 
 | | Cryo-EM structure of Arabidopsis phytochrome A. | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | Authors: | Ma, L, Zhou, C, Wang, J, Guan, Z, Yin, P. | | Deposit date: | 2023-02-17 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Plant phytochrome A in the Pr state assembles as an asymmetric dimer.
Cell Res., 33, 2023
|
|
8HXS
 
 | | Small_spotted catshark CD8alpha | | Descriptor: | T-cell surface glycoprotein CD8 alpha chain | | Authors: | Wang, J, Zou, J. | | Deposit date: | 2023-01-05 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The first crystal structure of CD8 alpha alpha from a cartilaginous fish.
Front Immunol, 14, 2023
|
|
6J5W
 
 | | Ligand-triggered allosteric ADP release primes a plant NLR complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Disease resistance RPP13-like protein 4, RKS1 | | Authors: | Wang, J.Z, Wang, J, Hu, M.J, Wang, H.W, Zhou, J.M, Chai, J.J. | | Deposit date: | 2019-01-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ligand-triggered allosteric ADP release primes a plant NLR complex.
Science, 364, 2019
|
|
6J6I
 
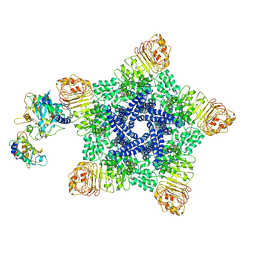 | | Reconstitution and structure of a plant NLR resistosome conferring immunity | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Disease resistance RPP13-like protein 4, Probable serine/threonine-protein kinase PBL2, ... | | Authors: | Wang, J.Z, Wang, J, Hu, M.J, Wang, H.W, Zhou, J.M, Chai, J.J. | | Deposit date: | 2019-01-15 | | Release date: | 2019-03-20 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Reconstitution and structure of a plant NLR resistosome conferring immunity.
Science, 364, 2019
|
|
6J5T
 
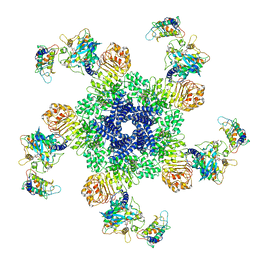 | | Reconstitution and structure of a plant NLR resistosome conferring immunity | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Disease resistance RPP13-like protein 4, Probable serine/threonine-protein kinase PBL2, ... | | Authors: | Wang, J.Z, Wang, J, Hu, M.J, Wang, H.W, Zhou, J.M, Chai, J.J. | | Deposit date: | 2019-01-12 | | Release date: | 2019-03-20 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Reconstitution and structure of a plant NLR resistosome conferring immunity.
Science, 364, 2019
|
|
6J5U
 
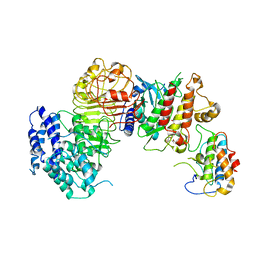 | | Ligand-triggered allosteric ADP release primes a plant NLR complex | | Descriptor: | Disease resistance RPP13-like protein 4, Probable serine/threonine-protein kinase PBL2, Protein kinase superfamily protein, ... | | Authors: | Wang, J.Z, Wang, J, Meijuan, H, Wang, H.W, Zhou, J.M, Chai, J.J. | | Deposit date: | 2019-01-12 | | Release date: | 2019-04-03 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ligand-triggered allosteric ADP release primes a plant NLR complex.
Science, 364, 2019
|
|
6J5V
 
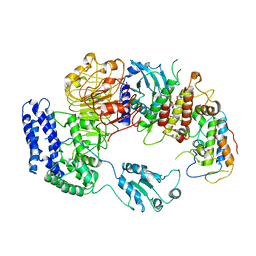 | | Ligand-triggered allosteric ADP release primes a plant NLR complex | | Descriptor: | Disease resistance RPP13-like protein 4, Probable serine/threonine-protein kinase PBL2, Protein kinase superfamily protein, ... | | Authors: | Wang, J.Z, Wang, J, Hu, M.J, Wang, H.W, Zhou, J.M, Chai, J.J. | | Deposit date: | 2019-01-12 | | Release date: | 2019-04-03 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Ligand-triggered allosteric ADP release primes a plant NLR complex.
Science, 364, 2019
|
|
3CFO
 
 | | Triple Mutant APO structure | | Descriptor: | DNA polymerase, GUANOSINE, SULFATE ION | | Authors: | Wang, J, Klimenko, D, Wang, M, Steitz, T.A, Konigsberg, W.H. | | Deposit date: | 2008-03-04 | | Release date: | 2009-03-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into base selectivity from the structures
of an RB69 DNA Polymerase triple mutant
To be Published
|
|
1VSC
 
 | | VCAM-1 | | Descriptor: | VASCULAR CELL ADHESION MOLECULE-1 | | Authors: | Wang, J, Stehle, T, Osborn, L. | | Deposit date: | 1995-04-27 | | Release date: | 1996-06-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of an N-terminal two-domain fragment of vascular cell adhesion molecule 1 (VCAM-1): a cyclic peptide based on the domain 1 C-D loop can inhibit VCAM-1-alpha 4 integrin interaction.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
3CFP
 
 | | Structure of the replicating complex of a POL Alpha family DNA Polymerase, ternary complex 1 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*DAP*DCP*DAP*DGP*DGP*DTP*DAP*DAP*DGP*DCP*DAP*DGP*DTP*DCP*DCP*DGP*DCP*DG)-3'), ... | | Authors: | Wang, J, Klimenko, D, Wang, M, Steitz, T.A, Konigsberg, W.H. | | Deposit date: | 2008-03-04 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into base selectivity from the structures
of an RB69 DNA Polymerase triple mutant
To be Published
|
|
3CFR
 
 | | Structure of the replicating complex of a POL Alpha family DNA Polymerase, ternary complex 2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*DGP*DCP*DGP*DGP*DAP*DCP*DTP*DGP*DCP*DTP*DTP*DAP*(DOC))-3'), ... | | Authors: | Wang, J, Klimenko, D, Wang, M, Steitz, T.A, Konigsberg, W.H. | | Deposit date: | 2008-03-04 | | Release date: | 2009-03-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into base selectivity from the structures
of an RB69 DNA Polymerase triple mutant
To be Published
|
|
