6E68
 
 | | NAMPT co-crystal with inhibitor compound 2 | | Descriptor: | (2E)-N-{4-[1-(3-aminobenzene-1-carbonyl)piperidin-4-yl]butyl}-3-(pyridin-3-yl)prop-2-enamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Waight, A.B, Neumann, C.S. | | Deposit date: | 2018-07-24 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | NAMPT co-crystal with inhibitor compound 2
to be published
|
|
6EZW
 
 | | Crystal structure of a llama VHH antibody BCD090-M2 against human ErbB3 in space group C2 | | Descriptor: | VHH antibody BCD090-M2 | | Authors: | Eliseev, I.E, Yudenko, A.N, Vysochinskaya, V.V, Svirina, A.A, Evstratyeva, A.V, Drozhzhachih, M.S, Krendeleva, E.A, Vladimirova, A.K, Nemankin, T.A, Ekimova, V.M, Ulitin, A.B, Lomovskaya, M.I, Yakovlev, P.A, Moiseenko, F.V, Chakchir, O.B. | | Deposit date: | 2017-11-16 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59842849 Å) | | Cite: | Crystal structures of a llama VHH antibody BCD090-M2 targeting human ErbB3 receptor.
F1000Res, 7, 2018
|
|
1C7T
 
 | | BETA-N-ACETYLHEXOSAMINIDASE MUTANT E540D COMPLEXED WITH DI-N ACETYL-D-GLUCOSAMINE (CHITOBIASE) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-N-ACETYLHEXOSAMINIDASE, SULFATE ION | | Authors: | Prag, G, Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K, Oppenheim, A.B. | | Deposit date: | 2000-03-17 | | Release date: | 2000-09-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of chitobiase mutants complexed with the substrate Di-N-acetyl-d-glucosamine: the catalytic role of the conserved acidic pair, aspartate 539 and glutamate 540.
J.Mol.Biol., 300, 2000
|
|
6F0D
 
 | | Crystal structure of a llama VHH antibody BCD090-M2 against human ErbB3 in space group P1 with cadmium ions | | Descriptor: | CADMIUM ION, VHH antibody BCD090-M2 | | Authors: | Eliseev, I.E, Yudenko, A.N, Vysochinskaya, V.V, Svirina, A.A, Evstratyeva, A.V, Drozhzhachih, M.S, Krendeleva, E.A, Vladimirova, A.K, Nemankin, T.A, Ekimova, V.M, Ulitin, A.B, Lomovskaya, M.I, Yakovlev, P.A, Moiseenko, F.V, Chakchir, O.B. | | Deposit date: | 2017-11-19 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.90000749 Å) | | Cite: | Crystal structures of a llama VHH antibody BCD090-M2 targeting human ErbB3 receptor.
F1000Res, 7, 2018
|
|
1CBV
 
 | | AN AUTOANTIBODY TO SINGLE-STRANDED DNA: COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF THE UNLIGANDED FAB AND A DEOXYNUCLEOTIDE-FAB COMPLEX | | Descriptor: | DNA (5'-D(*TP*TP*T)-3'), PROTEIN (FAB (BV04-01) AUTOANTIBODY-HEAVY CHAIN), PROTEIN (FAB (BV04-01) AUTOANTIBODY-LIGHT CHAIN) | | Authors: | Herron, J.N, He, X.M, Ballard, D.W, Blier, P.R, Pace, P.E, Bothwell, A.L.M, Voss Junior, E.W, Edmundson, A.B. | | Deposit date: | 1993-03-16 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | An autoantibody to single-stranded DNA: comparison of the three-dimensional structures of the unliganded Fab and a deoxynucleotide-Fab complex.
Proteins, 11, 1991
|
|
1BIZ
 
 | | HIV-1 INTEGRASE CORE DOMAIN | | Descriptor: | CACODYLATE ION, HIV-1 INTEGRASE | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-21 | | Release date: | 1998-08-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
6DZN
 
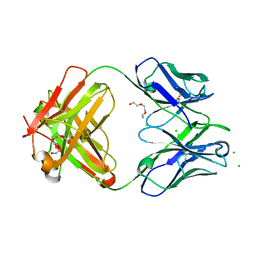 | | Pan-ebolavirus human antibody ADI-15878 Fab | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, Antibody ADI-15878, heavy chain, ... | | Authors: | Murin, C.D, Ward, A.B, Bruhn, J.F, Stanfield, R. | | Deposit date: | 2018-07-05 | | Release date: | 2018-09-12 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Pan-Ebolavirus Neutralization by an Antibody Targeting the Glycoprotein Fusion Loop.
Cell Rep, 24, 2018
|
|
1A8J
 
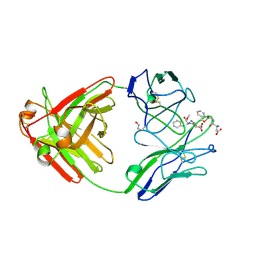 | |
1BIU
 
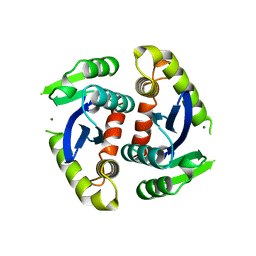 | | HIV-1 INTEGRASE CORE DOMAIN COMPLEXED WITH MG++ | | Descriptor: | HIV-1 INTEGRASE, MAGNESIUM ION | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-19 | | Release date: | 1998-08-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
8SWW
 
 | | BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody IF3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NHP polyclonal antibody IF3 Heavy Chain, ... | | Authors: | Pratap, P.P, Antansijevic, A, Ward, A.B. | | Deposit date: | 2023-05-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Priming antibody responses to the fusion peptide in rhesus macaques.
Npj Vaccines, 9, 2024
|
|
8SX3
 
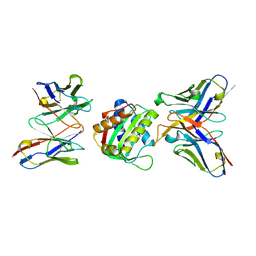 | | 10E8-GT10.2 immunogen in complex with human Fab 10E8 and mouse Fab W6-10 | | Descriptor: | 10E8 Fab heavy chain, 10E8 light chain, 10E8-GT10.2 immunogen, ... | | Authors: | Huang, J, Ozorowski, G, Ward, A.B. | | Deposit date: | 2023-05-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Vaccination induces broadly neutralizing antibody precursors to HIV gp41.
Nat.Immunol., 25, 2024
|
|
8SWV
 
 | | BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody IF1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IF1 Heavy Chain, ... | | Authors: | Pratap, P.P, Antansijevic, A, Ozorowski, G, Ward, A.B. | | Deposit date: | 2023-05-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Priming antibody responses to the fusion peptide in rhesus macaques.
Npj Vaccines, 9, 2024
|
|
8SWX
 
 | | BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody Base4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Base4 Heavy Chain, ... | | Authors: | Pratap, P.P, Antansijevic, A, Ward, A.B. | | Deposit date: | 2023-05-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Priming antibody responses to the fusion peptide in rhesus macaques.
Npj Vaccines, 9, 2024
|
|
8SW4
 
 | | BG505 GT1.1 SOSIP in complex with NHP Fabs 21N13, 21M20 and RM20A3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 21M20 heavy chain variable region, ... | | Authors: | Ozorowski, G, Torres, J.L, Zhang, S, Ward, A.B. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Neutralizing antibodies induced in non-human primates by germline targeting Env trimer
To Be Published
|
|
1W9S
 
 | |
1W9W
 
 | |
1W9T
 
 | | Structure of a beta-1,3-glucan binding CBM6 from Bacillus halodurans in complex with xylobiose | | Descriptor: | BH0236 PROTEIN, SODIUM ION, alpha-D-xylopyranose, ... | | Authors: | Boraston, A.B, van Bueren, A.L. | | Deposit date: | 2004-10-18 | | Release date: | 2004-11-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Family 6 Carbohydrate Binding Modules Recognize the Non-Reducing End of Beta-1,3-Linked Glucans by Presenting a Unique Ligand Binding Surface
J.Biol.Chem., 280, 2005
|
|
1YK8
 
 | | Cathepsin K complexed with a cyanamide-based inhibitor | | Descriptor: | Cathepsin K, TERT-BUTYL 2-CYANO-2-METHYLHYDRAZINECARBOXYLATE | | Authors: | Barrett, D.G, Deaton, D.N, Hassell, A.M, McFadyen, R.B, Miller, A.B, Miller, L.R, Payne, J.A, Shewchuk, L.M, Willard, D.H, Wright, L.L. | | Deposit date: | 2005-01-17 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Acyclic cyanamide-based inhibitors of cathepsin K.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1YK7
 
 | | Cathepsin K complexed with a cyanopyrrolidine inhibitor | | Descriptor: | Cathepsin K, N2-[(BENZYLOXY)CARBONYL]-N1-[(3S)-1-CYANOPYRROLIDIN-3-YL]-L-LEUCINAMIDE | | Authors: | Barrett, D.G, Deaton, D.N, Hassell, A.M, McFadyen, R.B, Miller, A.B, Miller, L.R, Shewchuk, L.M, Tavares, F.X, Willard, D.H, Wright, L.L. | | Deposit date: | 2005-01-17 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel and potent cyclic cyanamide-based cathepsin K inhibitors.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1WBH
 
 | |
1WA3
 
 | | Mechanism of the Class I KDPG aldolase | | Descriptor: | 2-KETO-3-DEOXY-6-PHOSPHOGLUCONATE ALDOLASE, PYRUVIC ACID, SULFATE ION | | Authors: | Fullerton, S.W.B, Griffiths, J.S, Merkel, A.B, Wymer, N.J, Hutchins, M.J, Fierke, C.A, Toone, E.J, Naismith, J.H. | | Deposit date: | 2004-10-22 | | Release date: | 2005-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of the Class I Kdpg Aldolase.
Bioorg.Med.Chem., 14, 2006
|
|
1WAU
 
 | |
1XVX
 
 | | Crystal Structure of iron-loaded Yersinia enterocolitica YfuA | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, FE (III) ION, ... | | Authors: | Shouldice, S.R, McRee, D.E, Dougan, D.R, Tari, L.W, Schryvers, A.B. | | Deposit date: | 2004-10-28 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Novel Anion-independent Iron Coordination by Members of a Third Class of Bacterial Periplasmic Ferric Ion-binding Proteins
J.Biol.Chem., 280, 2005
|
|
1YT7
 
 | | Cathepsin K complexed with a constrained ketoamide inhibitor | | Descriptor: | (1R)-2,2-DIMETHYL-1-({5-[4-(TRIFLUOROMETHYL)PHENYL]-1,3,4-OXADIAZOL-2-YL}METHYL)PROPYL (1S)-1-{OXO[(2-OXO-1,3-OXAZOLIDIN-3-YL)AMINO]ACETYL}PENTYLCARBAMATE, Cathepsin K, SULFATE ION | | Authors: | Barrett, D.G, Boncek, V.M, Catalano, J.G, Deaton, D.N, Hassell, A.M, Jurgensen, C.H, Long, S.T, McFadyen, R.B, Miller, A.B, Miller, L.R, Payne, J.A, Ray, J.A, Samano, V, Shewchuk, L.M, Tavares, F.X, Wells-Knecht, K.J, Willard, D.H, Wright, L.L, Zhou, H.Q. | | Deposit date: | 2005-02-10 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | P(2)-P(3) conformationally constrained ketoamide-based inhibitors of cathepsin K.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1XWR
 
 | | Crystal structure of the coliphage lambda transcription activator protein CII | | Descriptor: | ISOPROPYL ALCOHOL, Regulatory protein CII | | Authors: | Datta, A.B, Panjikar, S, Weiss, M.S, Chakrabarti, P, Parrack, P. | | Deposit date: | 2004-11-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure of {lambda} CII: Implications for recognition of direct-repeat DNA by an unusual tetrameric organization
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
