4KFS
 
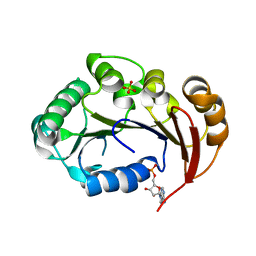 | | Structure of the genome packaging NTPase B204 from Sulfolobus turreted icosahedral virus 2 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CITRATE ANION, Genome packaging NTPase B204, ... | | Authors: | Happonen, L.J, Oksanen, E, Kajander, T, Goldman, A, Butcher, S. | | Deposit date: | 2013-04-27 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.946 Å) | | Cite: | The Structure of the NTPase That Powers DNA Packaging into Sulfolobus Turreted Icosahedral Virus 2.
J.Virol., 87, 2013
|
|
3M2L
 
 | |
2IV8
 
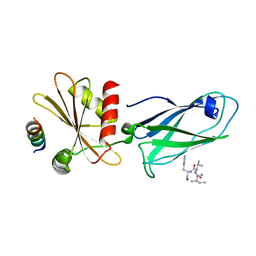 | |
2P58
 
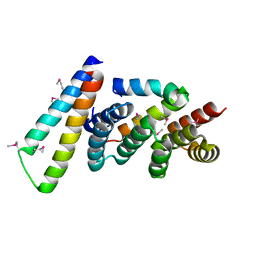 | | Structure of the Yersinia pestis Type III secretion system needle protein YscF in complex with its chaperones YscE/YscG | | Descriptor: | Putative type III secretion protein YscE, Putative type III secretion protein YscF, Putative type III secretion protein YscG | | Authors: | Sun, P, Austin, B.P, Tropea, J.E, Waugh, D.S. | | Deposit date: | 2007-03-14 | | Release date: | 2008-03-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the Yersinia pestis type III secretion system needle protein YscF in complex with its heterodimeric chaperone YscE/YscG.
J.Mol.Biol., 377, 2008
|
|
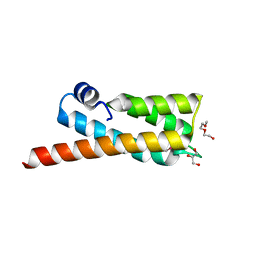
2OOC
 
 | |
4KFU
 
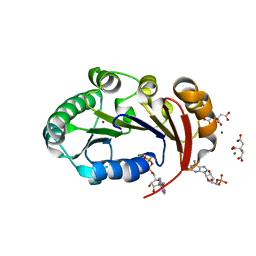 | | Structure of the genome packaging NTPase B204 from Sulfolobus turreted icosahedral virus 2 in complex with AMPPCP | | Descriptor: | CITRATE ANION, Genome packaging NTPase B204, MAGNESIUM ION, ... | | Authors: | Happonen, L.J, Oksanen, E, Kajander, T, Goldman, A, Butcher, S. | | Deposit date: | 2013-04-27 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | The Structure of the NTPase That Powers DNA Packaging into Sulfolobus Turreted Icosahedral Virus 2.
J.Virol., 87, 2013
|
|
3M3R
 
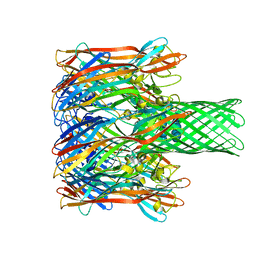 | |
5T8Q
 
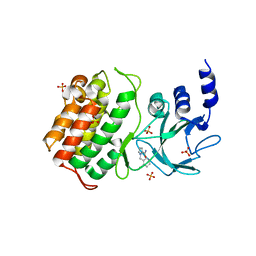 | |
2EVR
 
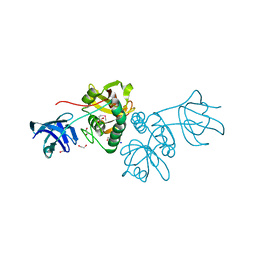 | |
5T8O
 
 | | Crystal structure of murine NF-kappaB inducing kinase (NIK) bound to Imidazobenzoxepin Compound 3 | | Descriptor: | 10-(3-methyl-3-oxidanyl-but-1-ynyl)-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepine-2-carboxamide, Mitogen-activated protein kinase kinase kinase 14, SULFATE ION | | Authors: | Smith, M.A, McEwan, P, Hymowitz, S.G. | | Deposit date: | 2016-09-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure-Based Design of Tricyclic NF-kappa B Inducing Kinase (NIK) Inhibitors That Have High Selectivity over Phosphoinositide-3-kinase (PI3K).
J. Med. Chem., 60, 2017
|
|
5T8P
 
 | | Crystal structure of murine NF-kappaB inducing kinase (NIK) bound to benzoxepin compound 2 | | Descriptor: | 6,7-dihydrothieno[4,5]oxepino[1,2-~{c}]pyridine-2-carboxamide, Mitogen-activated protein kinase kinase kinase 14, SULFATE ION | | Authors: | Smith, M.A, McEwan, P.A. | | Deposit date: | 2016-09-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure-Based Design of Tricyclic NF-kappa B Inducing Kinase (NIK) Inhibitors That Have High Selectivity over Phosphoinositide-3-kinase (PI3K).
J. Med. Chem., 60, 2017
|
|
1CBK
 
 | | 7,8-DIHYDRO-6-HYDROXYMETHYLPTERIN-PYROPHOSPHOKINASE FROM HAEMOPHILUS INFLUENZAE | | Descriptor: | 7,8-DIHYDRO-7,7-DIMETHYL-6-HYDROXYPTERIN, PROTEIN (7,8-DIHYDRO-6-HYDROXYMETHYLPTERIN-PYROPHOSPHOKINASE), SULFATE ION | | Authors: | Hennig, M, D'Arcy, A, Dale, G, Oefner, C. | | Deposit date: | 1999-02-26 | | Release date: | 2000-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The structure and function of the 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase from Haemophilus influenzae.
J.Mol.Biol., 287, 1999
|
|
4FYI
 
 | | Crystal structure of rcl with 6-cyclopentyl-AMP | | Descriptor: | Deoxyribonucleoside 5'-monophosphate N-glycosidase, N-cyclopentyladenosine 5'-(dihydrogen phosphate) | | Authors: | Labesse, G, Padilla, A, Kaminski, P.A. | | Deposit date: | 2012-07-04 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of the oncoprotein Rcl bound to three nucleotide analogues.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4KFT
 
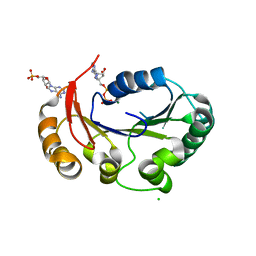 | | Structure of the genome packaging NTPase B204 from Sulfolobus turreted icosahedral virus 2 in complex with ATP-gammaS | | Descriptor: | CHLORIDE ION, CITRATE ANION, Genome packaging NTPase B204, ... | | Authors: | Happonen, L.J, Oksanen, E, Kajander, T, Goldman, A, Butcher, S. | | Deposit date: | 2013-04-27 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.241 Å) | | Cite: | The Structure of the NTPase That Powers DNA Packaging into Sulfolobus Turreted Icosahedral Virus 2.
J.Virol., 87, 2013
|
|
2HBW
 
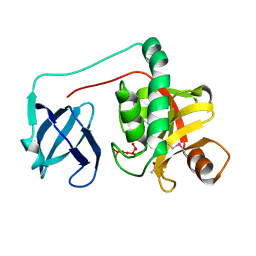 | |
1ODF
 
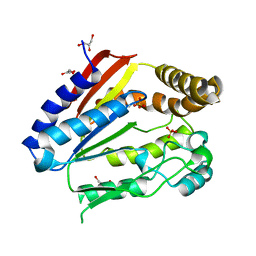 | | Structure of YGR205w protein. | | Descriptor: | GLYCEROL, HYPOTHETICAL 33.3 KDA PROTEIN IN ADE3-SER2 INTERGENIC REGION, SULFATE ION | | Authors: | Li De La Sierra-Gallay, I, Van Tilbeurgh, H. | | Deposit date: | 2003-02-19 | | Release date: | 2003-12-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Ygr205W Protein from Saccharomyces Cerevisiae: Close Structural Resemblance to E.Coli Pantothenate Kinase
Proteins: Struct.,Funct., Genet., 54, 2004
|
|
2MJE
 
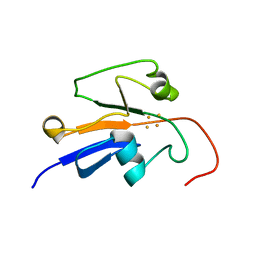 | | Reduced Yeast Adrenodoxin Homolog 1 | | Descriptor: | Adrenodoxin homolog, mitochondrial, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Gallo, A, Banci, L. | | Deposit date: | 2014-01-07 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Functional reconstitution of mitochondrial Fe/S cluster synthesis on Isu1 reveals the involvement of ferredoxin.
Nat Commun, 5, 2014
|
|
4XL1
 
 | | Complex of Notch1 (EGF11-13) bound to Delta-like 4 (N-EGF1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Luca, V.C, Jude, K.M, Garcia, K.C. | | Deposit date: | 2015-01-13 | | Release date: | 2015-03-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural biology. Structural basis for Notch1 engagement of Delta-like 4.
Science, 347, 2015
|
|
4XLW
 
 | | Complex of Notch1 (EGF11-13) bound to Delta-like 4 (N-EGF2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Delta-like protein, ... | | Authors: | Luca, V.C, Jude, K.M, Garcia, K.C. | | Deposit date: | 2015-01-13 | | Release date: | 2015-03-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Structural biology. Structural basis for Notch1 engagement of Delta-like 4.
Science, 347, 2015
|
|
2QI4
 
 | | Crystal structure of protease inhibitor, MIT-2-AD93 in complex with wild type HIV-1 protease | | Descriptor: | ACETATE ION, N-[(1S,2R)-3-{(1,3-BENZOTHIAZOL-6-YLSULFONYL)[(2S)-2-METHYLBUTYL]AMINO}-1-BENZYL-2-HYDROXYPROPYL]-3-HYDROXYBENZAMIDE, PHOSPHATE ION, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
2QI1
 
 | | Crystal structure of protease inhibitor, MIT-1-KK81 in complex with wild type HIV-1 protease | | Descriptor: | N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{[(3-METHOXYPHENYL)SULFONYL](2-THIENYLMETHYL)AMINO}PROPYL]-3,4-DIHYDROXYBENZAMIDE, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
2QHZ
 
 | | Crystal structure of protease inhibitor, MIT-1-AC87 in complex with wild type HIV-1 protease | | Descriptor: | (2E)-N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{(2-THIENYLMETHYL)[(2,4,5-TRIFLUOROPHENYL)SULFONYL]AMINO}PROPYL]-4,4,4-TRIFLUORO-3-METHYLBUT-2-ENAMIDE, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
2QI0
 
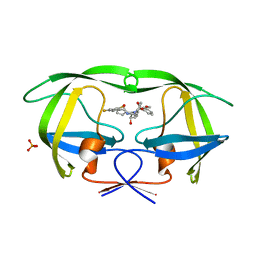 | |
2QHY
 
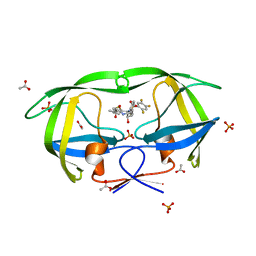 | | Crystal Structure of protease inhibitor, MIT-1-AC86 in complex with wild type HIV-1 protease | | Descriptor: | ACETATE ION, N~2~-ACETYL-N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{(2-THIENYLMETHYL)[(2,4,5-TRIFLUOROPHENYL)SULFONYL]AMINO}PROPYL]-L-ALANINAMIDE, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
7F24
 
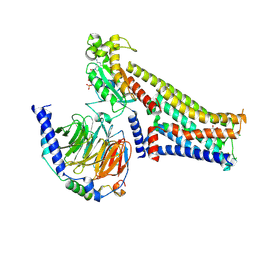 | |
