8UQH
 
 | | X-ray crystal structure of PRMT4 bound to compound YD-1130 | | Descriptor: | 5'-S-(2-{[(3-bromophenyl)methyl]amino}ethyl)-5'-thioadenosine, Histone-arginine methyltransferase CARM1 | | Authors: | Bush, M, Noinaj, N, Deng, Y, Huang, R. | | Deposit date: | 2023-10-23 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | An Adenosine Analogue Library Reveals Insights into Active Sites of Protein Arginine Methyltransferases and Enables the Discovery of a Selective PRMT4 Inhibitor.
J.Med.Chem., 67, 2024
|
|
8HTX
 
 | | Crystal structure of BANP in complex with methylated DNA | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*(5CM)P*GP*CP*GP*AP*GP*AP*G)-3'), Protein BANP | | Authors: | Zhang, J, Min, J, Liu, K. | | Deposit date: | 2022-12-21 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into DNA recognition by the BEN domain of the transcription factor BANP.
J.Biol.Chem., 299, 2023
|
|
8I7X
 
 | | Crystal structure of human ClpP in complex with ZG36 | | Descriptor: | (6S,9aS)-N-[(4-bromophenyl)methyl]-6-[(2S)-butan-2-yl]-8-[(4-methoxynaphthalen-1-yl)methyl]-4,7-bis(oxidanylidene)-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Wang, P.Y, Gan, J.H, Yang, C.-G. | | Deposit date: | 2023-02-02 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Assessment of the structure-activity relationship and antileukemic activity of diacylpyramide compounds as human ClpP agonists.
Eur.J.Med.Chem., 258, 2023
|
|
9KNI
 
 | | Structural complex of FTO bound with 12j | | Descriptor: | 2-OXOGLUTARIC ACID, 3-[[2,6-bis(chloranyl)-4-(3,5-dimethyl-1H-pyrazol-4-yl)phenyl]amino]thiophene-2-carboxylic acid, Alpha-ketoglutarate-dependent dioxygenase FTO | | Authors: | Yang, C.G, Gan, J.H. | | Deposit date: | 2024-11-19 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Development of 3-arylaminothiophenic-2-carboxylic acid derivatives as new FTO inhibitors showing potent antileukemia activities.
Eur.J.Med.Chem., 289, 2025
|
|
8K9D
 
 | | Structure of human Caprin-2 HR1 domain | | Descriptor: | Caprin-2 | | Authors: | Song, X.M. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-21 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insights into the Caprin-2 HR1 domain in canonical Wnt signaling.
J.Biol.Chem., 300, 2024
|
|
5HHV
 
 | |
5HHX
 
 | |
9IRV
 
 | | MultiBody Refinement of dimeric DARPin and its bound GFP on a symmetric scaffold | | Descriptor: | DARPin, Green fluorescent protein | | Authors: | Lu, X, Yan, M, Zhang, H.M, Hao, Q. | | Deposit date: | 2024-07-16 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | A large, general and modular DARPin-apoferritin scaffold enables the visualization of small proteins by cryo-EM.
Iucrj, 12, 2025
|
|
9IVP
 
 | | 24-mer DARPin-apoferritin scaffold in complex with the maltose binding protein | | Descriptor: | DARPin,Ferritin heavy chain, N-terminally processed, Maltodextrin-binding protein | | Authors: | Lu, X, Yan, M, Zhang, H.M, Hao, Q. | | Deposit date: | 2024-07-24 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A large, general and modular DARPin-apoferritin scaffold enables the visualization of small proteins by cryo-EM.
Iucrj, 12, 2025
|
|
9JMA
 
 | | Crystal structure of glycosyltransferase AvpGT | | Descriptor: | GalNAc(5)-diNAcBac-PP-undecaprenol beta-1,3-glucosyltransferase, MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Li, J.H, Wang, Z.Q, Zhang, Z.Y, Chen, W.Q. | | Deposit date: | 2024-09-20 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Efficient Synthesis of Glycodiversified Nucleoside Analogues by a Thermophilic Promiscuous Glycosyltransferase
Acs Catalysis, 15, 2025
|
|
9JN3
 
 | | Crystal structure of AvpGT in complex with Ara-A | | Descriptor: | 2-(6-AMINO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, GalNAc(5)-diNAcBac-PP-undecaprenol beta-1,3-glucosyltransferase, MAGNESIUM ION, ... | | Authors: | Li, J.H, Wang, Z.Q, Zhang, Z.Y, Chen, W.Q. | | Deposit date: | 2024-09-22 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Efficient Synthesis of Glycodiversified Nucleoside Analogues by a Thermophilic Promiscuous Glycosyltransferase
Acs Catalysis, 15, 2025
|
|
7YC9
 
 | | Co-crystal structure of BTK kinase domain with inhibitor | | Descriptor: | (7~{S})-2-(4-bromanyl-3,5-dimethoxy-phenyl)-7-(1-propanoylpiperidin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, 1,2-ETHANEDIOL, Tyrosine-protein kinase BTK | | Authors: | Zhou, X. | | Deposit date: | 2022-07-01 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of BGB-8035, a Highly Selective Covalent Inhibitor of Bruton's Tyrosine Kinase for B-Cell Malignancies and Autoimmune Diseases.
J.Med.Chem., 66, 2023
|
|
8JO4
 
 | | Cryo-EM structure of a Legionella effector complexed with actin and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Zhou, X.T, Wang, X.F, Tan, J.X, Zhu, Y.Q. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Legionella effector LnaB is a phosphoryl AMPylase that impairs phosphosignalling.
Nature, 631, 2024
|
|
8JO3
 
 | | Cryo-EM structure of a Legionella effector complexed with actin and AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Zhou, X.T, Wang, X.F, Tan, J.X, Zhu, Y.Q. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Legionella effector LnaB is a phosphoryl AMPylase that impairs phosphosignalling.
Nature, 631, 2024
|
|
7Y4T
 
 | | Crystal structure of cMET kinase domain bound by compound 9I | | Descriptor: | 2-[2-[3-(1-methylpyrazol-4-yl)quinolin-6-yl]ethyl]-6-(3-nitrophenyl)pyridazin-3-one, Hepatocyte growth factor receptor | | Authors: | Qu, L.Z, Chen, Y.H. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Discovery of D6808, a Highly Selective and Potent Macrocyclic c-Met Inhibitor for Gastric Cancer Harboring MET Gene Alteration Treatment.
J.Med.Chem., 65, 2022
|
|
7Y4U
 
 | | Crystal structure of cMET kinase domain bound by compound 9Y | | Descriptor: | Hepatocyte growth factor receptor, ~{N}-methyl-4-[1-[2-[3-(1-methylpyrazol-4-yl)quinolin-6-yl]ethyl]-6-oxidanylidene-pyridazin-3-yl]-2-(trifluoromethyl)benzamide | | Authors: | Qu, L.Z, Chen, Y.H. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of D6808, a Highly Selective and Potent Macrocyclic c-Met Inhibitor for Gastric Cancer Harboring MET Gene Alteration Treatment.
J.Med.Chem., 65, 2022
|
|
8KH8
 
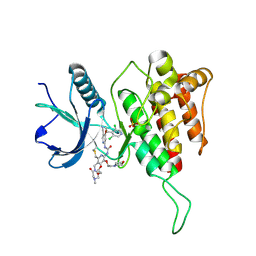 | | Crystal structure of FGFR4(V550L) kinase domain with 8z | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-3-[6-methanoyl-5-[(4-methyl-2-oxidanylidene-piperazin-1-yl)methyl]-3-(2-morpholin-4-ylethoxy)pyridin-2-yl]urea, Fibroblast growth factor receptor 4, ... | | Authors: | Lin, Q.M, Chen, X.J, Chen, Y.H. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of 6-Formylpyridyl Urea Derivatives as Potent Reversible-Covalent Fibroblast Growth Factor Receptor 4 Inhibitors with Improved Anti-Hepatocellular Carcinoma Activity.
J.Med.Chem., 67, 2024
|
|
8KH6
 
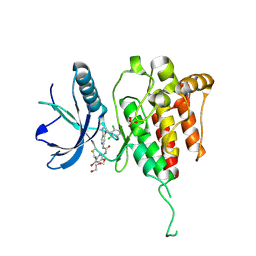 | | Crystal structure of FGFR4 kinase domain with 8r | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-3-[5-bromanyl-6-(hydroxymethyl)-3-methoxy-pyridin-2-yl]urea, Fibroblast growth factor receptor 4, ... | | Authors: | Lin, Q.M, Chen, X.J, Chen, Y.H. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery of 6-Formylpyridyl Urea Derivatives as Potent Reversible-Covalent Fibroblast Growth Factor Receptor 4 Inhibitors with Improved Anti-Hepatocellular Carcinoma Activity.
J.Med.Chem., 67, 2024
|
|
8KH9
 
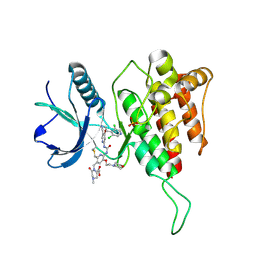 | | Crystal structure of FGFR4(V550M) kinase domain with 8z | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-3-[6-methanoyl-5-[(4-methyl-2-oxidanylidene-piperazin-1-yl)methyl]-3-(2-morpholin-4-ylethoxy)pyridin-2-yl]urea, Fibroblast growth factor receptor 4, ... | | Authors: | Lin, Q.M, Chen, X.J, Chen, Y.H. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Discovery of 6-Formylpyridyl Urea Derivatives as Potent Reversible-Covalent Fibroblast Growth Factor Receptor 4 Inhibitors with Improved Anti-Hepatocellular Carcinoma Activity.
J.Med.Chem., 67, 2024
|
|
8KH7
 
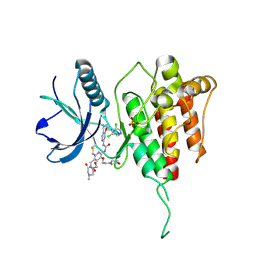 | | Crystal structure of FGFR4 kinase domain with 8zc | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-3-[6-methanoyl-5-[(4-methyl-2-oxidanylidene-piperazin-1-yl)methyl]-3-(2-morpholin-4-ylethoxy)pyridin-2-yl]urea, Fibroblast growth factor receptor 4, ... | | Authors: | Lin, Q.M, Chen, X.J, Chen, Y.H. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of 6-Formylpyridyl Urea Derivatives as Potent Reversible-Covalent Fibroblast Growth Factor Receptor 4 Inhibitors with Improved Anti-Hepatocellular Carcinoma Activity.
J.Med.Chem., 67, 2024
|
|
7YBO
 
 | | Crystal structure of FGFR4 kinase domain with 10z | | Descriptor: | Fibroblast growth factor receptor 4, SULFATE ION, ~{N}-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-6-bromanyl-5-(hydroxymethyl)-1-(2-morpholin-4-ylethyl)pyrrolo[3,2-b]pyridine-3-carboxamide | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2022-06-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7YC3
 
 | | Crystal structure of FGFR4 kinase domain with 10t | | Descriptor: | 6-bromanyl-~{N}-[5-cyano-4-(2-methoxyethylamino)pyridin-2-yl]-5-methanoyl-1-(2-morpholin-4-ylethyl)pyrrolo[3,2-b]pyridine-3-carboxamide, Fibroblast growth factor receptor 4, GLYCEROL, ... | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2022-06-30 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.987 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7YC1
 
 | | Crystal structure of FGFR4 kinase domain with 10d | | Descriptor: | Fibroblast growth factor receptor 4, GLYCEROL, SULFATE ION, ... | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2022-06-30 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.535 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7YBP
 
 | | Crystal structure of FGFR4(V550L) kinase domain with 10z | | Descriptor: | Fibroblast growth factor receptor 4, SULFATE ION, ~{N}-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-6-bromanyl-5-(hydroxymethyl)-1-(2-morpholin-4-ylethyl)pyrrolo[3,2-b]pyridine-3-carboxamide | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2022-06-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7YBX
 
 | | Crystal structure of FGFR4(V550M) kinase domain with 10z | | Descriptor: | Fibroblast growth factor receptor 4, SULFATE ION, ~{N}-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-6-bromanyl-5-(hydroxymethyl)-1-(2-morpholin-4-ylethyl)pyrrolo[3,2-b]pyridine-3-carboxamide | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2022-06-30 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.233 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
