7EGD
 
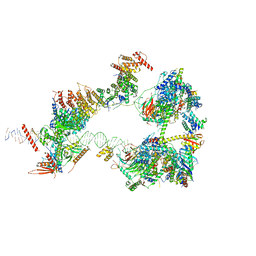 | | SCP promoter-bound TFIID-TFIIA in initial TBP-loading state | | Descriptor: | DNA (72-MER), TATA-box-binding protein, Transcription initiation factor IIA subunit 1, ... | | Authors: | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (6.75 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EGJ
 
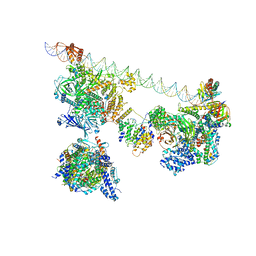 | | SCP promoter-bound TFIID-TFIIA in post TBP-loading state | | Descriptor: | DNA (74-MER), TATA-box-binding protein, Transcription initiation factor IIA subunit 1, ... | | Authors: | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (8.64 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EG8
 
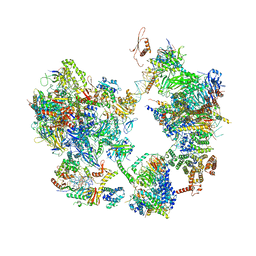 | | TFIID-based core PIC on PUMA promoter | | Descriptor: | DNA (85-MER), DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Chen, X, Qi, Y, Hou, H, Wang, X, Wu, Z, Li, J, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-05 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EGE
 
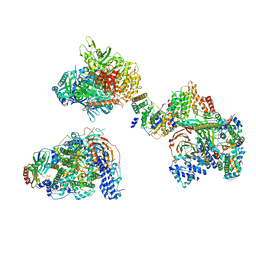 | | TFIID in canonical conformation | | Descriptor: | TATA-box-binding protein, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 10, ... | | Authors: | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EGC
 
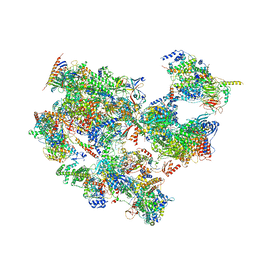 | | p53-bound TFIID-based holo PIC on HDM2 promoter | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Wu, Z, Hou, H, Qi, Y, Wang, X, Li, J, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7Y8R
 
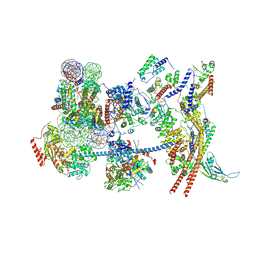 | | The nucleosome-bound human PBAF complex | | Descriptor: | ACTB protein (Fragment), ADENOSINE-5'-DIPHOSPHATE, AT-rich interactive domain-containing protein 2, ... | | Authors: | Wang, L, Yu, J, Yu, Z, Wang, Q, He, S, Xu, Y. | | Deposit date: | 2022-06-24 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of nucleosome-bound human PBAF complex.
Nat Commun, 13, 2022
|
|
7FJO
 
 | | Cryo-EM structure of South African (B.1.351) SARS-CoV-2 spike glycoprotein in complex with three T6 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
7FJN
 
 | | Cryo-EM structure of South African (B.1.351) SARS-CoV-2 spike glycoprotein in complex with two T6 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Envelope glycoprotein, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
7DD5
 
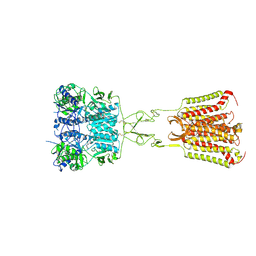 | | Structure of Calcium-Sensing Receptor in complex with NPS-2143 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-chloro-6-[(2R)-2-hydroxy-3-{[2-methyl-1-(naphthalen-2-yl)propan-2-yl]amino}propoxy]benzonitrile, ... | | Authors: | Wen, T.L, Yang, X, Shen, Y.Q. | | Deposit date: | 2020-10-27 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for activation and allosteric modulation of full-length calcium-sensing receptor.
Sci Adv, 7, 2021
|
|
7DD6
 
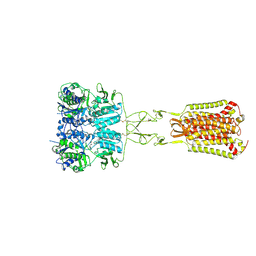 | | Structure of Ca2+/L-Trp-bonnd Calcium-Sensing Receptor in active state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wen, T.L, Yang, X, Shen, Y.Q. | | Deposit date: | 2020-10-27 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for activation and allosteric modulation of full-length calcium-sensing receptor.
Sci Adv, 7, 2021
|
|
7DD7
 
 | | Structure of Calcium-Sensing Receptor in complex with Evocalcet | | Descriptor: | 2-[4-[(3S)-3-[[(1R)-1-naphthalen-1-ylethyl]amino]pyrrolidin-1-yl]phenyl]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wen, T.L, Yang, X, Shen, Y.Q. | | Deposit date: | 2020-10-27 | | Release date: | 2021-06-16 | | Last modified: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for activation and allosteric modulation of full-length calcium-sensing receptor.
Sci Adv, 7, 2021
|
|
7EHL
 
 | | Cryo-EM structure of human ABCB8 transporter in nucleotide binding state | | Descriptor: | CHOLESTEROL, MAGNESIUM ION, Mitochondrial potassium channel ATP-binding subunit, ... | | Authors: | Li, S.J, Yang, X, Shen, Y.Q. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-05 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY | | Cite: | Cryo-EM structure of human ABCB8 transporter in nucleotide binding state.
Biochem.Biophys.Res.Commun., 557, 2021
|
|
7FCC
 
 | | IL-1RAcPb TIR domain | | Descriptor: | Isoform 4 of Interleukin-1 receptor accessory protein | | Authors: | Wang, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | Structural basis of the IL-1 receptor TIR domain-mediated IL-1 signaling
Iscience, 25, 2022
|
|
7FCJ
 
 | | Zebrafish SIGIRR TIR domain mutant - C299S | | Descriptor: | SIGIRR protein | | Authors: | Wang, X, Zhou, J. | | Deposit date: | 2021-07-15 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural basis of the IL-1 receptor TIR domain-mediated IL-1 signaling
Iscience, 25, 2022
|
|
7FCH
 
 | | IL-18Rbeta TIR domain | | Descriptor: | Interleukin-18 receptor accessory protein | | Authors: | Wang, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.883 Å) | | Cite: | Structural basis of the IL-1 receptor TIR domain-mediated IL-1 signaling
Iscience, 25, 2022
|
|
7FD3
 
 | | IL-1RAPL2 TIR domain | | Descriptor: | X-linked interleukin-1 receptor accessory protein-like 2 | | Authors: | Wang, X, Zhou, J. | | Deposit date: | 2021-07-15 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis of the IL-1 receptor TIR domain-mediated IL-1 signaling
Iscience, 25, 2022
|
|
6IIK
 
 | | USP14 catalytic domain with IU1 | | Descriptor: | 1-[1-(4-fluorophenyl)-2,5-dimethyl-1H-pyrrol-3-yl]-2-(pyrrolidin-1-yl)ethan-1-one, Ubiquitin carboxyl-terminal hydrolase 14 | | Authors: | Mei, Z.Q, Wang, Y.W, He, W, Wang, F. | | Deposit date: | 2018-10-06 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Small molecule inhibitors reveal allosteric regulation of USP14 via steric blockade.
Cell Res., 28, 2018
|
|
6IIN
 
 | | USP14 catalytic domain with IU1-248 | | Descriptor: | 4-{3-[(4-hydroxypiperidin-1-yl)acetyl]-2,5-dimethyl-1H-pyrrol-1-yl}benzonitrile, Ubiquitin carboxyl-terminal hydrolase 14 | | Authors: | Mei, Z.Q, Wang, J.W, Wang, F, Wang, Y.W. | | Deposit date: | 2018-10-07 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Small molecule inhibitors reveal allosteric regulation of USP14 via steric blockade.
Cell Res., 28, 2018
|
|
6IIL
 
 | | USP14 catalytic domain bind to IU1-47 | | Descriptor: | 1-[1-(4-chlorophenyl)-2,5-dimethyl-1H-pyrrol-3-yl]-2-(piperidin-1-yl)ethan-1-one, Ubiquitin carboxyl-terminal hydrolase 14 | | Authors: | Mei, Z.Q, Wang, Y.W, Wang, F, Wang, J.W, He, W, Ding, S, Li, J.W. | | Deposit date: | 2018-10-07 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Small molecule inhibitors reveal allosteric regulation of USP14 via steric blockade.
Cell Res., 28, 2018
|
|
6JKV
 
 | | PppA, a key regulatory component of T6SS in Pseudomonas aeruginosa | | Descriptor: | MANGANESE (II) ION, PppA | | Authors: | Wang, T, Liu, L, Wu, Y, Li, D. | | Deposit date: | 2019-03-02 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PppA from Pseudomonas aeruginosa, a key regulatory component of type VI secretion systems.
Biochem.Biophys.Res.Commun., 516, 2019
|
|
6IIM
 
 | | USP14 catalytic domain with IU1-206 | | Descriptor: | 1-[1-(4-chlorophenyl)-2,5-dimethyl-1H-pyrrol-3-yl]-2-(4-hydroxypiperidin-1-yl)ethan-1-one, Ubiquitin carboxyl-terminal hydrolase 14 | | Authors: | Mei, Z.Q, Wang, J.W, Wang, F, Wang, Y.W. | | Deposit date: | 2018-10-07 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Small molecule inhibitors reveal allosteric regulation of USP14 via steric blockade.
Cell Res., 28, 2018
|
|
4L07
 
 | | Crystal structure of the maleamate amidase Ami from Pseudomonas putida S16 | | Descriptor: | GLYCEROL, Hydrolase, isochorismatase family, ... | | Authors: | Chen, D.D, Lu, Y, Zhang, Z, Wu, G, Xu, P. | | Deposit date: | 2013-05-30 | | Release date: | 2014-07-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the specific recognition of N-heterocycle biodenitrogenation-derived substrates by microbial amide hydrolases.
Mol.Microbiol., 91, 2014
|
|
4K7F
 
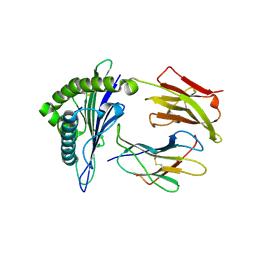 | | Newly identified epitope V60 from HBV core protein complexed with HLA-A*0201 | | Descriptor: | Beta-2-microglobulin, Core protein, HLA class I histocompatibility antigen, ... | | Authors: | Meng, S.D, Zhang, Y, Wu, Y, Qi, J.X. | | Deposit date: | 2013-04-17 | | Release date: | 2013-06-05 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The L60V variation in hepatitis B virus core protein elicits new epitope-specific cytotoxic T lymphocytes and enhances viral replication.
J.Virol., 87, 2013
|
|
4L08
 
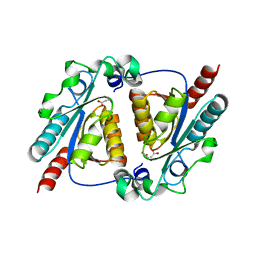 | | Crystal structure of the maleamate amidase Ami(C149A) in complex with maleate from Pseudomonas putida S16 | | Descriptor: | Hydrolase, isochorismatase family, MALEIC ACID | | Authors: | Chen, D.D, Lu, Y, Zhang, Z, Wu, G, Xu, P. | | Deposit date: | 2013-05-31 | | Release date: | 2014-07-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural insights into the specific recognition of N-heterocycle biodenitrogenation-derived substrates by microbial amide hydrolases.
Mol.Microbiol., 91, 2014
|
|
8J8Q
 
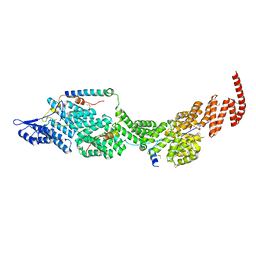 | | Structure of the four-component Paf1 complex from Saccharomyces eubayanus | | Descriptor: | CDC73-like protein, CTR9-like protein, PAF1-like protein, ... | | Authors: | Wang, Z, Qin, Y, Zhou, Y, Cao, Y. | | Deposit date: | 2023-05-02 | | Release date: | 2023-05-24 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural Basis of the Transcriptional Elongation Factor Paf1 Core Complex from Saccharomyces eubayanus .
Int J Mol Sci, 24, 2023
|
|
