4MZW
 
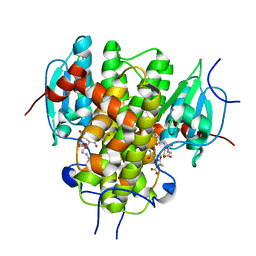 | | CRYSTAL STRUCTURE OF NU-CLASS GLUTATHIONE TRANSFERASE YGHU FROM Streptococcus sanguinis SK36, COMPLEX WITH GLUTATHIONE DISULFIDE, TARGET EFI-507286 | | Descriptor: | ACETATE ION, Glutathione S-Transferase, OXIDIZED GLUTATHIONE DISULFIDE | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase Yghu (Target Efi-507286)
To be Published
|
|
4JYX
 
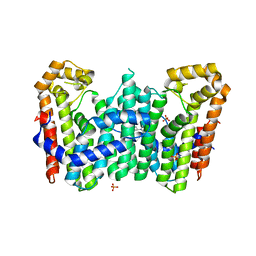 | | Crystal structure of polyprenyl synthase PATL_3739 (TARGET EFI-509195) FROM PSEUDOALTEROMONAS ATLANTICA, COMPLEX WITH INORGANIC PHOSPHATE AND AN UNKNOWN LIGAND | | Descriptor: | PHOSPHATE ION, Trans-hexaprenyltranstransferase, UNKNOWN LIGAND | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-04-01 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Isoprenoid Synthase Patl_3739 from Pseudoalteromonas Atlantica
To be Published
|
|
2RNE
 
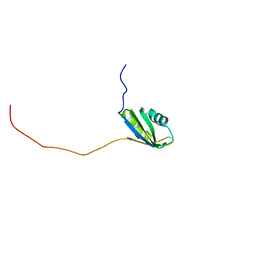 | | Solution structure of the second RNA recognition motif (RRM) of TIA-1 | | Descriptor: | Tia1 protein | | Authors: | Takahashi, M, Kuwasako, K, Abe, C, Tsuda, K, Inoue, M, Terada, T, Shirouzu, M, Kobayashi, N, Kigawa, T, Taguchi, S, Guntert, P, Hayashizaki, Y, Tanaka, A, Muto, Y, Yokoyama, S. | | Deposit date: | 2007-12-19 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second RNA recognition motif (RRM) domain of murine T cell intracellular antigen-1 (TIA-1) and its RNA recognition mode
Biochemistry, 47, 2008
|
|
1UFN
 
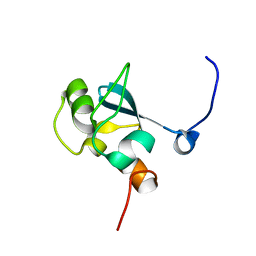 | | Solution structure of the SAND domain of the putative nuclear protein homolog (5830484A20Rik) | | Descriptor: | putative nuclear protein homolog 5830484A20Rik | | Authors: | Tochio, N, Kobayashi, N, Koshiba, S, Kigawa, T, Inoue, M, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Matsuo, Y, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-02 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SAND domain of the putative nuclear protein homolog (5830484A20Rik)
To be Published
|
|
1UFZ
 
 | | Solution structure of HBS1-like domain in hypothetical protein BAB28515 | | Descriptor: | Hypothetical protein BAB28515 | | Authors: | He, F, Muto, Y, Hayami, N, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-11 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of HBS1-like domain in hypothetical protein BAB28515
To be Published
|
|
1UG8
 
 | | NMR structure of the R3H domain from Poly(A)-specific Ribonuclease | | Descriptor: | Poly(A)-specific Ribonuclease | | Authors: | Nagata, T, Muto, Y, Hayami, N, Uda, H, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-15 | | Release date: | 2004-08-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the R3H domain from Poly(A)-specific Ribonuclease
To be Published
|
|
1UE9
 
 | | Solution structure of the fourth SH3 domain of human intersectin 2 (KIAA1256) | | Descriptor: | Intersectin 2 | | Authors: | Tochio, N, Kigawa, T, Koshiba, S, Kobayashi, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-09 | | Release date: | 2003-11-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fourth SH3 domain of human intersectin 2 (KIAA1256)
To be Published
|
|
1UFX
 
 | | Solution structure of the third PDZ domain of human KIAA1526 protein | | Descriptor: | KIAA1526 protein | | Authors: | Tochio, N, Kobayashi, N, Koshiba, S, Kigawa, T, Inoue, M, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Matsuo, Y, Ohara, O, Nagase, T, Kikuno, R, Nakayama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-10 | | Release date: | 2003-12-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third PDZ domain of human KIAA1526 protein
To be Published
|
|
1WGP
 
 | | Solution structure of the cNMP-binding domain from Arabidopsis thaliana cyclic nucleotide-regulated ion channel | | Descriptor: | Probable cyclic nucleotide-gated ion channel 6 | | Authors: | Chikayama, E, Nameki, N, Kigawa, T, Koshiba, S, Inoue, M, Tomizawa, T, Kobayashi, N, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cNMP-binding domain from Arabidopsis thaliana cyclic nucleotide-regulated ion channel
To be Published
|
|
2RSX
 
 | | Solution structure of IseA, an inhibitor protein of DL-endopeptidases from Bacillus subtilis | | Descriptor: | Uncharacterized protein yoeB | | Authors: | Arai, R, Li, H, Tochio, N, Fukui, S, Kobayashi, N, Kitaura, C, Watanabe, S, Kigawa, T, Sekiguchi, J. | | Deposit date: | 2012-08-09 | | Release date: | 2012-10-31 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of IseA, an Inhibitor Protein of DL-Endopeptidases from Bacillus subtilis, Reveals a Novel Fold with a Characteristic Inhibitory Loop
J.Biol.Chem., 287, 2012
|
|
1UJY
 
 | | Solution structure of SH3 domain in Rac/Cdc42 guanine nucleotide exchange factor(GEF) 6 | | Descriptor: | Rho guanine nucleotide exchange factor 6 | | Authors: | He, F, Muto, Y, Uda, H, Koshiba, S, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Ohara, O, Nagase, T, Kikuno, R, Nagayama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-08-12 | | Release date: | 2004-02-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SH3 domain in Rac/Cdc42 guanine nucleotide exchange factor(GEF) 6
To be Published
|
|
3V4B
 
 | | Crystal structure of an enolase from the soil bacterium Cellvibrio japonicus (TARGET EFI-502161) with bound MG and L-tartrate | | Descriptor: | CHLORIDE ION, L(+)-TARTARIC ACID, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Seidel, R.D, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Al Obaidi, N, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of an enolase from the soil bacterium Cellvibrio japonicus (TARGET EFI-502161) with bound MG and l-tartrate
to be published
|
|
3FAL
 
 | | humanRXR alpha & mouse LXR alpha complexed with Retenoic acid and GSK2186 | | Descriptor: | 2-{4-[butyl(3-chloro-4,5-dimethoxybenzyl)amino]phenyl}-1,1,1,3,3,3-hexafluoropropan-2-ol, Oxysterols receptor LXR-alpha, RETINOIC ACID, ... | | Authors: | Chao, E.Y, Caravella, J.A, Watson, M.A, Campobasso, N, Ghisletti, S, Billin, A.N, Galardi, C, Willson, T.M, Zuercher, W.J, Collins, J.L. | | Deposit date: | 2008-11-17 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure-guided design of N-phenyl tertiary amines as transrepression-selective liver X receptor modulators with anti-inflammatory activity.
J.Med.Chem., 51, 2008
|
|
3FC6
 
 | | hRXRalpha & mLXRalpha with an indole Pharmacophore, SB786875 | | Descriptor: | Nr1h3 protein, RETINOIC ACID, Retinoic acid receptor RXR-alpha, ... | | Authors: | Washburn, D.G, Hoang, T.H, Campobasso, N, Smallwood, A, Parks, D.J, Webb, C.L, Frank, K, Nord, M, Duraiswami, C, Evans, C, Jaye, M, Thompson, S.K. | | Deposit date: | 2008-11-21 | | Release date: | 2009-02-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.063 Å) | | Cite: | Synthesis and SAR of potent LXR agonists containing an indole pharmacophore.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3PYY
 
 | | Discovery and Characterization of a Cell-Permeable, Small-molecule c-Abl Kinase Activator that Binds to the Myristoyl Binding Site | | Descriptor: | (5R)-5-[3-(4-fluorophenyl)-1-phenyl-1H-pyrazol-4-yl]imidazolidine-2,4-dione, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, GLYCEROL, ... | | Authors: | Yang, J, Campobasso, N, Biju, M.P, Fisher, K, Pan, X.Q, Cottom, J, Galbraith, S, Ho, T, Zhang, H, Hong, X, Ward, P, Hofmann, G, Siegfried, B. | | Deposit date: | 2010-12-13 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and Characterization of a Cell-Permeable, Small-Molecule c-Abl Kinase Activator that Binds to the Myristoyl Binding Site.
Chem.Biol., 18, 2011
|
|
2RUJ
 
 | | Solution structure of MTSL spin-labeled Schizosaccharomyces pombe Sin1 CRIM domain | | Descriptor: | Stress-activated map kinase-interacting protein 1 | | Authors: | Furuita, K, Kataoka, S, Sugiki, T, Kobayashi, N, Ikegami, T, Shiozaki, K, Fujiwara, T, Kojima, C. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-29 | | Method: | SOLUTION NMR | | Cite: | Utilization of paramagnetic relaxation enhancements for high-resolution NMR structure determination of a soluble loop-rich protein with sparse NOE distance restraints
J.Biomol.Nmr, 61, 2015
|
|
2RQ4
 
 | | Refinement of RNA binding domain 3 in CUG triplet repeat RNA-binding protein 1 | | Descriptor: | CUG-BP- and ETR-3-like factor 1 | | Authors: | Tsuda, K, Kuwasako, K, Takahashi, M, Someya, T, Inoue, M, Terada, T, Kobayashi, N, Shirouzu, M, Kigawa, T, Guntert, P, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-01-19 | | Release date: | 2009-08-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3
Nucleic Acids Res., 2009
|
|
1UFW
 
 | | Solution structure of RNP domain in Synaptojanin 2 | | Descriptor: | Synaptojanin 2 | | Authors: | He, F, Muto, Y, Ushikoshi, R, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Ohara, O, Nagase, T, Kikuno, R, Nakayama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-10 | | Release date: | 2003-12-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RNP domain in Synaptojanin 2
To be Published
|
|
2RRL
 
 | | Solution structure of the C-terminal domain of the FliK | | Descriptor: | Flagellar hook-length control protein | | Authors: | Mizuno, S, Tate, S, Kobayashi, N, Amida, H. | | Deposit date: | 2011-01-02 | | Release date: | 2011-05-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of FliK, the trigger for the switch of substrate specificity in the flagellar type III secretion apparatus
J.Mol.Biol., 409, 2011
|
|
2RU3
 
 | | Solution structure of c.elegans SUP-12 RRM in complex with RNA | | Descriptor: | Protein SUP-12, isoform a, RNA (5'-R(*GP*UP*GP*UP*GP*C)-3') | | Authors: | Takahashi, M, Kuwasako, K, Unzai, S, Tsuda, K, Yoshikawa, S, He, F, Kobayashi, N, Guntert, P, Shirouzu, M, Ito, T, Tanaka, A, Yokoyama, S, Hagiwara, M, Kuroyanagi, H, Muto, Y. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | RBFOX and SUP-12 sandwich a G base to cooperatively regulate tissue-specific splicing
Nat.Struct.Mol.Biol., 21, 2014
|
|
2RUG
 
 | | Refined solution structure of the first RNA recognition motif domain in CPEB3 | | Descriptor: | Cytoplasmic polyadenylation element-binding protein 3 | | Authors: | Tsuda, K, Kuwasako, K, Nagata, T, Takahashi, M, Kigawa, T, Kobayashi, N, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2014-04-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Novel RNA recognition motif domain in the cytoplasmic polyadenylation element binding protein 3.
Proteins, 82, 2014
|
|
1UG0
 
 | | Solution structure of SURP domain in BAB30904 | | Descriptor: | splicing factor 4 | | Authors: | He, F, Muto, Y, Ushikoshi, R, Koshiba, S, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-11 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SURP domain in BAB30904
To be Published
|
|
1UG1
 
 | | SH3 domain of Hypothetical protein BAA76854.1 | | Descriptor: | KIAA1010 protein | | Authors: | Nagata, T, Muto, Y, Kamewari, Y, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Ohara, O, Nagase, T, Kikuno, R, Nakayama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-11 | | Release date: | 2003-12-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of SH3 domain of Hypothetical protein BAA76854.1
To be Published
|
|
1UHC
 
 | | Solution Structure of RSGI RUH-002, a SH3 Domain of KIAA1010 protein [Homo sapiens] | | Descriptor: | KIAA1010 protein | | Authors: | Abe, T, Hirota, H, Kobayashi, N, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-28 | | Release date: | 2003-12-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of RSGI RUH-002, a SH3 Domain of KIAA1010 protein [Homo sapiens]
To be Published
|
|
1UIL
 
 | | Double-stranded RNA-binding motif of Hypothetical protein BAB28848 | | Descriptor: | Double-stranded RNA-binding motif | | Authors: | Nagata, T, Muto, Y, Hayashi, F, Hamana, H, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-17 | | Release date: | 2004-11-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of Double-stranded RNA-binding motif of Hypothetical protein BAB28848
To be Published
|
|
