6SSN
 
 | | RNASE 3/1 version3 | | Descriptor: | GLYCEROL, PHOSPHATE ION, RNase 3/1 version3 | | Authors: | Fernandez-Millan, P, Prats-Ejarque, G, Vazquez-Monteagudo, S, Boix, E. | | Deposit date: | 2019-09-08 | | Release date: | 2021-10-06 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | Exploring the RNase A scaffold to combine catalytic and antimicrobial activities. Structural characterization of RNase 3/1 chimeras.
Front Mol Biosci, 9, 2022
|
|
6SSO
 
 | | EDN mutant L45H | | Descriptor: | ACETATE ION, Non-secretory ribonuclease | | Authors: | Fernandez-Millan, P, Prats-Ejarque, G, Vazquez-Monteagudo, S, Boix, E. | | Deposit date: | 2019-09-08 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.211 Å) | | Cite: | Structural and functional characterization of new family enzymes derivates from human RNase 1 and 3 with antimicrobial and ribonuclease activity
To Be Published
|
|
6TNS
 
 | | PI3K delta in complex with 2methoxyN[2methoxy5(7{[(2R)4(oxetan3 yl)morpholin2yl]methoxy}1,3dihydro2 benzofuran5yl)pyridin3yl]ethane1 sulfonamide | | Descriptor: | 2-methoxy-~{N}-[2-methoxy-5-[7-[[(2~{R})-4-(oxetan-3-yl)morpholin-2-yl]methoxy]-1,3-dihydro-2-benzofuran-5-yl]pyridin-3-yl]ethanesulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Convery, M.A, Rowland, P, Henley, Z.A, Barton, N, Down, K. | | Deposit date: | 2019-12-10 | | Release date: | 2020-01-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Optimization of Orally Bioavailable PI3K delta Inhibitors and Identification of Vps34 as a Key Selectivity Target.
J.Med.Chem., 63, 2020
|
|
2JAI
 
 | | DDAH1 complexed with citrulline | | Descriptor: | CITRULLINE, NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE 1 | | Authors: | Murray-Rust, J, O'Hara, B.P, Rossiter, S, Leiper, J.M, Vallance, P, McDonald, N.Q. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Disruption of methylarginine metabolism impairs vascular homeostasis.
Nat. Med., 13, 2007
|
|
5LXL
 
 | | NMR structure of the N-terminal domain of the Bacteriophage T5 decoration protein pb10 | | Descriptor: | Decoration protein | | Authors: | Vernhes, E, Gilquin, B, Cuniasse, P, Boulanger, P, Zinn-Justin, S. | | Deposit date: | 2016-09-22 | | Release date: | 2017-04-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid.
Sci Rep, 7, 2017
|
|
2JAJ
 
 | | DDAH1 complexed with L-257 | | Descriptor: | NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE 1, N~5~-{IMINO[(2-METHOXYETHYL)AMINO]METHYL}-L-ORNITHINE | | Authors: | Murray-Rust, J, O'Hara, B.P, Rossiter, S, Leiper, J.M, Vallance, P, McDonald, N.Q. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Disruption of methylarginine metabolism impairs vascular homeostasis.
Nat. Med., 13, 2007
|
|
5LXK
 
 | | NMR structure of the C-terminal domain of the Bacteriophage T5 decoration protein pb10. | | Descriptor: | Decoration protein | | Authors: | Vernhes, E, Gilquin, B, Cuniasse, P, Boulanger, P, Zinn-Justin, S. | | Deposit date: | 2016-09-22 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid.
Sci Rep, 7, 2017
|
|
2GVE
 
 | | Time-of-Flight Neutron Diffraction Structure of D-Xylose Isomerase | | Descriptor: | COBALT (II) ION, Xylose isomerase | | Authors: | Katz, A.K, Li, X, Carrell, H.L, Hanson, B.L, Langan, P, Coates, L, Schoenborn, B.P, Glusker, J.P, Bunick, G.J. | | Deposit date: | 2006-05-02 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Locating active-site hydrogen atoms in D-xylose isomerase: Time-of-flight neutron diffraction.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2O2U
 
 | | Crystal structure of human JNK3 complexed with N-(3-cyano-4,5,6,7-tetrahydro-1-benzothien-2-yl)-2-fluorobenzamide | | Descriptor: | Mitogen-activated protein kinase 10, N-(3-cyano-4,5,6,7-tetrahydro-1-benzothien-2-yl)-2-fluorobenzamide | | Authors: | Somers, D, Rowland, P. | | Deposit date: | 2006-11-30 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | N-(3-Cyano-4,5,6,7-tetrahydro-1-benzothien-2-yl)amides as potent, selective, inhibitors of JNK2 and JNK3.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2PKY
 
 | | The Effect of Deuteration on Protein Structure A High Resolution Comparison of Hydrogenous and Perdeuterated Haloalkane Dehalogenase | | Descriptor: | Haloalkane dehalogenase | | Authors: | Liu, X, Hanson, L, Langan, P, Viola, R.E. | | Deposit date: | 2007-04-18 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The effect of deuteration on protein structure: a high-resolution comparison of hydrogenous and perdeuterated haloalkane dehalogenase.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
5FN0
 
 | | Crystal structure of Pseudomonas fluorescens kynurenine-3- monooxygenase (KMO) in complex with GSK180 | | Descriptor: | 3-(5,6-DICHLORO-2-OXOBENZO[D]OXAZOL-3(2H)-YL)PROPANOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, KYNURENINE 3-MONOOXYGENASE | | Authors: | Mole, D.J, Webster, S.P, Uings, I, Zheng, X, Binnie, M, Wilson, K, Hutchinson, J.P, Mirguet, O, Walker, A, Beaufils, B, Ancellin, N, Trottet, L, Beneton, V, Mowat, C.G, Wilkinson, M, Rowland, P, Haslam, C, McBride, A, Homer, N.Z.M, Baily, J.E, Sharp, M.G.F, Garden, O.J, Hughes, J, Howie, S.E.M, Holmes, D, Liddle, J, Iredale, J.P. | | Deposit date: | 2015-11-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Kynurenine-3-Monooxygenase Inhibition Prevents Multiple Organ Failure in Rodent Models of Acute Pancreatitis.
Nat.Med. (N.Y.), 22, 2016
|
|
3NOQ
 
 | | Crystal Structure of C101S Isocyanide Hydratase from Pseudomonas fluorescens | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ThiJ/PfpI family protein | | Authors: | Lakshminarasimhan, M, Madzelan, P, Nan, R, Milkovic, N.M, Wilson, M.A. | | Deposit date: | 2010-06-25 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Evolution of New Enzymatic Function by Structural Modulation of Cysteine Reactivity in Pseudomonas fluorescens Isocyanide Hydratase.
J.Biol.Chem., 285, 2010
|
|
3NOR
 
 | | Crystal Structure of T102S Isocyanide Hydratase from Pseudomonas fluorescens | | Descriptor: | CITRIC ACID, ThiJ/PfpI family protein | | Authors: | Lakshminarasimhan, M, Madzelan, P, Nan, R, Milkovic, N.M, Wilson, M.A. | | Deposit date: | 2010-06-25 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of New Enzymatic Function by Structural Modulation of Cysteine Reactivity in Pseudomonas fluorescens Isocyanide Hydratase.
J.Biol.Chem., 285, 2010
|
|
3NOV
 
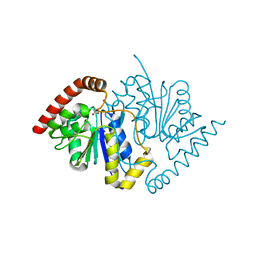 | | Crystal Structure of D17E Isocyanide Hydratase from Pseudomonas fluorescens | | Descriptor: | ACETATE ION, ThiJ/PfpI family protein | | Authors: | Lakshminarasimhan, M, Madzelan, P, Nan, R, Milkovic, N.M, Wilson, M.A. | | Deposit date: | 2010-06-25 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Evolution of New Enzymatic Function by Structural Modulation of Cysteine Reactivity in Pseudomonas fluorescens Isocyanide Hydratase.
J.Biol.Chem., 285, 2010
|
|
3NPZ
 
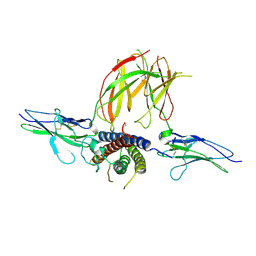 | | Prolactin Receptor (PRLR) Complexed with the Natural Hormone (PRL) | | Descriptor: | Prolactin, Prolactin receptor | | Authors: | Van Agthoven, J, England, P, Goffin, V, Broutin, I. | | Deposit date: | 2010-06-29 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural characterization of the stem-stem dimerization interface between prolactin receptor chains complexed with the natural hormone.
J.Mol.Biol., 404, 2010
|
|
1N5Y
 
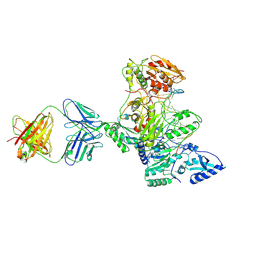 | | HIV-1 Reverse Transcriptase Crosslinked to Post-Translocation AZTMP-Terminated DNA (Complex P) | | Descriptor: | 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3', 5'-D(*AP*TP*GP*C*TP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3', MAGNESIUM ION, ... | | Authors: | Sarafianos, S.G, Clark Jr, A.D, Das, K, Tuske, S, Birktoft, J.J, Ilankumaran, P, Ramesha, A.R, Sayer, J.M, Jerina, D.M, Boyer, P.L, Hughes, S.H, Arnold, E. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-28 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of HIV-1 Reverse Transcriptase with Pre-Translocation and Post-Translocation AZTMP-Terminated DNA
Embo J., 21, 2002
|
|
3NOO
 
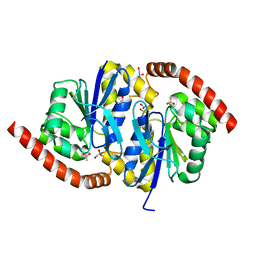 | | Crystal Structure of C101A Isocyanide Hydratase from Pseudomonas fluorescens | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ThiJ/PfpI family protein | | Authors: | Lakshminarasimhan, M, Madzelan, P, Nan, R, Milkovic, N.M, Wilson, M.A. | | Deposit date: | 2010-06-25 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Evolution of New Enzymatic Function by Structural Modulation of Cysteine Reactivity in Pseudomonas fluorescens Isocyanide Hydratase.
J.Biol.Chem., 285, 2010
|
|
3CWH
 
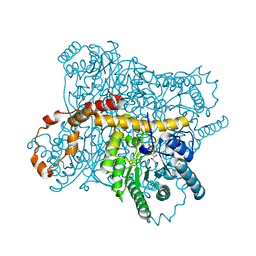 | | D-xylose Isomerase in complex with linear product, per-deuterated xylulose | | Descriptor: | D-XYLULOSE, HYDROXIDE ION, MAGNESIUM ION, ... | | Authors: | Kovalevsky, A.Y, Langan, P, Glusker, J.P. | | Deposit date: | 2008-04-21 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Hydrogen location in stages of an enzyme-catalyzed reaction: time-of-flight neutron structure of D-xylose isomerase with bound D-xylulose
Biochemistry, 47, 2008
|
|
3NON
 
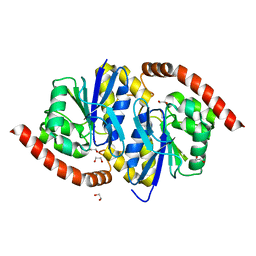 | | Crystal Structure of Isocyanide Hydratase from Pseudomonas fluorescens | | Descriptor: | 1,2-ETHANEDIOL, Isocyanide hydratase | | Authors: | Lakshminarasimhan, M, Madzelan, P, Nan, R, Milkovic, N.M, Wilson, M.A. | | Deposit date: | 2010-06-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Evolution of New Enzymatic Function by Structural Modulation of Cysteine Reactivity in Pseudomonas fluorescens Isocyanide Hydratase.
J.Biol.Chem., 285, 2010
|
|
3EW3
 
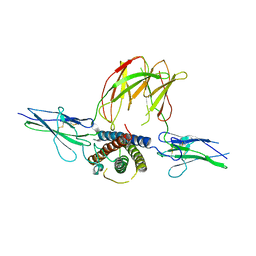 | | the 1:2 complex between a Nterminal elongated prolactin and the extra cellular domain of the rat prolactin receptor | | Descriptor: | Prolactin, Prolactin receptor | | Authors: | Broutin, I, Jomain, J.B, England, P, Goffin, V. | | Deposit date: | 2008-10-14 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of an affinity-matured prolactin complexed to its dimerized receptor reveals the topology of hormone binding site 2.
J.Biol.Chem., 285, 2010
|
|
3TQ6
 
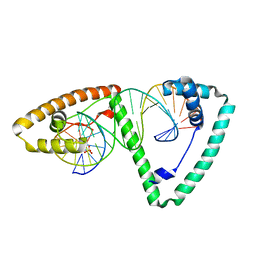 | | Crystal structure of human mitochondrial transcription factor A, TFAM or mtTFA, bound to the light strand promoter LSP | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA (5'-D(*GP*TP*TP*AP*GP*TP*TP*GP*GP*GP*GP*GP*GP*TP*GP*AP*CP*TP*GP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*CP*AP*GP*TP*CP*AP*CP*CP*CP*CP*CP*CP*AP*AP*CP*(BRU)P*AP*AP*C)-3'), ... | | Authors: | Rubio-Cosials, A, Sydow, J.F, Jimenez-Menendez, N, Fernandez-Millan, P, Montoya, J, Jacobs, H.T, Coll, M, Bernado, P, Sola, M. | | Deposit date: | 2011-09-09 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2YXP
 
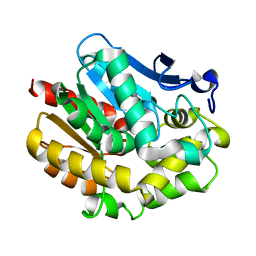 | | The Effect of Deuteration on Protein Structure A High Resolution Comparison of Hydrogenous and Perdeuterated Haloalkane Dehalogenase | | Descriptor: | Haloalkane dehalogenase | | Authors: | Liu, X, Hanson, L, Langan, P, Viola, R.E. | | Deposit date: | 2007-04-27 | | Release date: | 2007-09-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The effect of deuteration on protein structure: a high-resolution comparison of hydrogenous and perdeuterated haloalkane dehalogenase.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4HLL
 
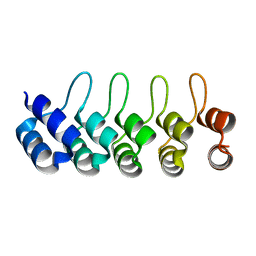 | | Crystal structure of Artificial ankyrin repeat protein_Ank(GAG)1D4 | | Descriptor: | Ankyrin(GAG)1D4 | | Authors: | Chuankhayan, P, Nangola, S, Minard, P, Boulanger, P, Hong, S.S, Tayapiwatana, C, Chen, C.-J. | | Deposit date: | 2012-10-17 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of Gag bioactive determinants specific to designed ankyrin and interfering in HIV-1 assembly
To be Published
|
|
4ZFH
 
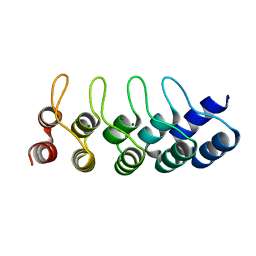 | | Crystal structure of Artificial ankyrin repeat protein_Ank(GAG)1D4 mutant -Y56A | | Descriptor: | Artificial ankyrin repeat protein_Ank(GAG)1D4 mutant -Y56A, MAGNESIUM ION | | Authors: | Chuankhayan, P, Saoin, S, Chupradit, K, Wisitponchai, T, Intachai, K, Kitidee, K, Nangola, S, Sanghiran, L.V, Hong, S.S, Boulanger, P, Tayapiwatana, C, Chen, C.J. | | Deposit date: | 2015-04-21 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of Artificial ankyrin repeat protein_Ank(GAG)1D4 mutant- Y56A
To Be Published
|
|
2VNN
 
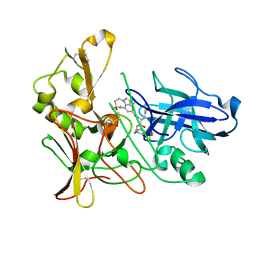 | | Human BACE-1 in complex with 7-ethyl-N-((1S,2R)-2-hydroxy-1-(phenylmethyl)-3-(((3-(trifluoromethyl)phenyl)methyl)amino)propyl)-1- methyl-3,4-dihydro-1H-(1,2,5)thiadiazepino(3,4,5-hi)indole-9- carboxamide 2,2-dioxide | | Descriptor: | BETA-SECRETASE 1, N-[(1S,2R)-1-benzyl-2-hydroxy-3-{[3-(trifluoromethyl)benzyl]amino}propyl]-7-ethyl-1-methyl-3,4-dihydro-1H-[1,2,5]thiadiazepino[3,4,5-hi]indole-9-carboxamide 2,2-dioxide | | Authors: | Charrier, N, Clarke, B, Cutler, L, Demont, E, Dingwall, C, Dunsdon, R, East, P, Hawkins, J, Howes, C, Hussain, I, Jeffrey, P, Maile, G, Matico, R, Mosley, J, Naylor, A, OBrien, A, Redshaw, S, Rowland, P, Soleil, V, Smith, K.J, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Wayne, G. | | Deposit date: | 2008-02-05 | | Release date: | 2008-05-20 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Second Generation of Hydroxyethylamine Bace-1 Inhibitors: Optimizing Potency and Oral Bioavailability.
J.Med.Chem., 51, 2008
|
|
