1IH0
 
 | | Structure of the C-domain of Human Cardiac Troponin C in Complex with Ca2+ Sensitizer EMD 57033 | | Descriptor: | 5-[1-(3,4-DIMETHOXY-BENZOYL)-1,2,3,4-TETRAHYDRO-QUINOLIN-6-YL]-6-METHYL-3,6-DIHYDRO-[1,3,4]THIADIAZIN-2-ONE, CALCIUM ION, TROPONIN C, ... | | Authors: | Wang, X, Li, M.X, Spyracopoulos, L, Beier, N, Chandra, M, Solaro, R.J, Sykes, B.D. | | Deposit date: | 2001-04-18 | | Release date: | 2001-10-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-domain of human cardiac troponin C in complex with the Ca2+ sensitizing drug EMD 57033.
J.Biol.Chem., 276, 2001
|
|
3TZD
 
 | | Crystal structure of the complex of Human Chromobox Homolog 3 (CBX3) | | Descriptor: | Chromobox protein homolog 3, Histone H1.4 | | Authors: | Amaya, M.F, Ravichandran, M, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-09-27 | | Release date: | 2012-03-07 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis of the chromodomain of Cbx3 bound to methylated peptides from histone h1 and G9a.
Plos One, 7, 2012
|
|
4ER8
 
 | | Structure of the REP associates tyrosine transposase bound to a REP hairpin | | Descriptor: | DNA (32-MER), NICKEL (II) ION, TnpArep for protein | | Authors: | Messing, S.A.J, Ton-Hoang, B, Hickman, A.B, Ghirlando, R, Chandler, M, Dyda, F. | | Deposit date: | 2012-04-19 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The processing of repetitive extragenic palindromes: the structure of a repetitive extragenic palindrome bound to its associated nuclease.
Nucleic Acids Res., 40, 2012
|
|
2K28
 
 | | Solution NMR structure of the chromo domain of the chromobox protein homolog 4 | | Descriptor: | E3 SUMO-protein ligase CBX4 | | Authors: | Kaustov, L, Lemak, A, Quyang, H, Fares, C, Gutmanas, A, Ravichandran, M, Loppnau, P, Bountra, C, Weigelt, J, Edwards, A.M, Min, J, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-27 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the chromo domain of the chromobox protein homolog 4.
To be Published
|
|
2K1B
 
 | | Solution NMR structure of the chromo domain of the chromobox protein homolog 7 | | Descriptor: | Chromobox protein homolog 7 | | Authors: | Kaustov, L, Lemak, A, Quyang, H, Gutmanas, A, Fares, C, Bountra, C, Weigelt, J, Loppnau, P, Ravichandran, M, Edwards, A.M, Min, J, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-25 | | Release date: | 2008-03-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the chromobox protein homolog 7.
To be Published
|
|
4EDU
 
 | | The MBT repeats of human SCML2 in a complex with histone H2A peptide | | Descriptor: | Histone H2A.J peptide, Sex comb on midleg-like protein 2 | | Authors: | Nady, N, Amaya, M.F, Tempel, W, Ravichandran, M, Arrowsmith, C.H. | | Deposit date: | 2012-03-27 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Histone recognition by human malignant brain tumor domains.
J.Mol.Biol., 423, 2012
|
|
8B3E
 
 | | Variant Surface Glycoprotein VSG397 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Variant surface glycoprotein 397 | | Authors: | Zeelen, J.P, Stebbins, C.E, Dakovic, S, Foti, K. | | Deposit date: | 2022-09-16 | | Release date: | 2023-01-25 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structural similarities between the metacyclic and bloodstream form variant surface glycoproteins of the African trypanosome.
Plos Negl Trop Dis, 17, 2023
|
|
5WOE
 
 | |
7NE3
 
 | | Human TET2 in complex with favourable DNA substrate. | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (5'-D(*AP*CP*AP*GP*GP*(5CM)P*GP*CP*CP*TP*G)-3'), ... | | Authors: | Rafalski, D, Bochtler, M. | | Deposit date: | 2021-02-03 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Pronounced sequence specificity of the TET enzyme catalytic domain guides its cellular function.
Sci Adv, 8, 2022
|
|
7NE6
 
 | | Human TET2 in complex with unfavourable DNA substrate. | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (5'-D(*AP*CP*AP*GP*GP*(5CM)P*GP*CP*CP*TP*G)-3'), ... | | Authors: | Rafalski, D, Bochtler, M. | | Deposit date: | 2021-02-03 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pronounced sequence specificity of the TET enzyme catalytic domain guides its cellular function.
Sci Adv, 8, 2022
|
|
4K3J
 
 | | Crystal structure of Onartuzumab Fab in complex with MET and HGF-beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor beta chain, ... | | Authors: | Ma, X, Starovasnik, M.A. | | Deposit date: | 2013-04-10 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Monovalent antibody design and mechanism of action of onartuzumab, a MET antagonist with anti-tumor activity as a therapeutic agent.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5KU3
 
 | | BRD4 bromodomain in complex with Cpd59 ((S)-1-(3-((2-fluoro-4-(1-methyl-1H-pyrazol-4-yl)phenyl)amino)-1-(tetrahydrofuran-3-yl)-6,7-dihydro-1H-pyrazolo[4,3-c]pyridin-5(4H)-yl)ethanone) | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-[[2-fluoranyl-4-(1-methylpyrazol-4-yl)phenyl]amino]-1-[(3~{S})-oxolan-3-yl]-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-5-yl]ethanone, Bromodomain-containing protein 4, ... | | Authors: | Murray, J.M, Huang, W. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Discovery of a Potent and Selective in Vivo Probe (GNE-272) for the Bromodomains of CBP/EP300.
J. Med. Chem., 59, 2016
|
|
4R8W
 
 | | Crystal structure of H7 hemagglutinin from A/Anhui/1/2013 in complex with a neutralizing antibody CT149 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of neutralizing antibody CT149, Hemagglutinin, ... | | Authors: | Wu, Y, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2014-09-03 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.795 Å) | | Cite: | A potent broad-spectrum protective human monoclonal antibody crosslinking two haemagglutinin monomers of influenza A virus
Nat Commun, 6, 2015
|
|
7O4A
 
 | |
7O49
 
 | |
7O4C
 
 | |
7RP3
 
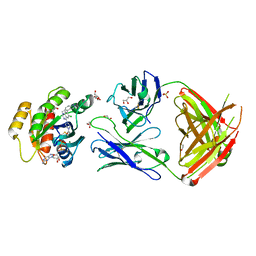 | | Crystal structure of GNE-1952 alkylated KRAS G12C in complex with 2H11 CLAMP | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[6-chloranyl-7-(5-methyl-1~{H}-indazol-4-yl)quinazolin-4-yl]piperazin-1-yl]propan-1-one, GLYCEROL, ... | | Authors: | Oh, A, Tam, C, Wang, W. | | Deposit date: | 2021-08-03 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformation-locking antibodies for the discovery and characterization of KRAS inhibitors.
Nat.Biotechnol., 40, 2022
|
|
7RP2
 
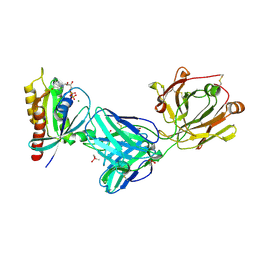 | | Crystal structure of Kas G12C in complex with 2H11 CLAMP | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, GTPase KRas, ... | | Authors: | Oh, A, Tam, C, Wang, W. | | Deposit date: | 2021-08-03 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformation-locking antibodies for the discovery and characterization of KRAS inhibitors.
Nat.Biotechnol., 40, 2022
|
|
7RP4
 
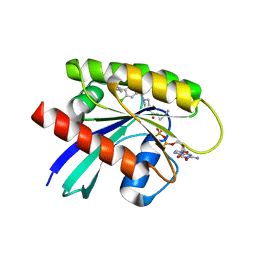 | | Crystal structure of KRAS G12C in complex with GNE-1952 | | Descriptor: | 1-[4-[6-chloranyl-7-(5-methyl-1~{H}-indazol-4-yl)quinazolin-4-yl]piperazin-1-yl]propan-1-one, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Oh, A, Tam, C, Wang, W. | | Deposit date: | 2021-08-03 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformation-locking antibodies for the discovery and characterization of KRAS inhibitors.
Nat.Biotechnol., 40, 2022
|
|
5KUC
 
 | | Crystal structure of trypsin activated Cry6Aa | | Descriptor: | Pesticidal crystal protein Cry6Aa | | Authors: | Kelker, M.S, Xu, X, Lee, M, Chan, M, Hung, S, Dementiev, K, Hey, T, Chikwana, V.M, Narva, K.E. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The pesticidal Cry6Aa toxin from Bacillus thuringiensis is structurally similar to HlyE-family alpha pore-forming toxins.
Bmc Biol., 14, 2016
|
|
5KUD
 
 | | Crystal structure of full length Cry6Aa | | Descriptor: | Pesticidal crystal protein Cry6Aa | | Authors: | Kelker, M.S, Xu, X, Lee, M, Chan, M, Hung, S, Dementiev, K, Chikwana, V.M, Hey, T, Narva, K. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The pesticidal Cry6Aa toxin from Bacillus thuringiensis is structurally similar to HlyE-family alpha pore-forming toxins.
Bmc Biol., 14, 2016
|
|
3ZG0
 
 | | Crystal structure of ceftaroline acyl-PBP2a from MRSA with non- covalently bound ceftaroline and muramic acid at allosteric site obtained by cocrystallization | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ceftaroline, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | How Allosteric Control of Staphylococcus Aureus Penicillin Binding Protein 2A Enables Methicillin Resistance and Physiological Function
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3ZFZ
 
 | | Crystal structure of ceftaroline acyl-PBP2a from MRSA with non- covalently bound ceftaroline and muramic acid at allosteric site obtained by soaking | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ceftaroline, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | How Allosteric Control of Staphylococcus Aureus Penicillin Binding Protein 2A Enables Methicillin Resistance and Physiological Function
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3ZG5
 
 | | Crystal structure of PBP2a from MRSA in complex with peptidoglycan analogue at allosteric | | Descriptor: | CADMIUM ION, CHLORIDE ION, PEPTIDOGLYCAN ANALOGUE, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2012-12-14 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | How Allosteric Control of Staphylococcus Aureus Penicillin Binding Protein 2A Enables Methicillin Resistance and Physiological Function
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5KTX
 
 | | CREBBP bromodomain in complex with Cpd59 ((S)-1-(3-((2-fluoro-4-(1-methyl-1H-pyrazol-4-yl)phenyl)amino)-1-(tetrahydrofuran-3-yl)-6,7-dihydro-1H-pyrazolo[4,3-c]pyridin-5(4H)-yl)ethanone) | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-[[2-fluoranyl-4-(1-methylpyrazol-4-yl)phenyl]amino]-1-[(3~{S})-oxolan-3-yl]-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-5-yl]ethanone, CREB-binding protein, ... | | Authors: | Murray, J.M, Noland, C. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Discovery of a Potent and Selective in Vivo Probe (GNE-272) for the Bromodomains of CBP/EP300.
J. Med. Chem., 59, 2016
|
|
