4N9J
 
 | | Crystal structure of the cryptic polo box domain of human Plk4 | | Descriptor: | CHLORIDE ION, SULFATE ION, Serine/threonine-protein kinase PLK4 | | Authors: | Ku, B, Kim, J.H, Lee, K.S, Kim, S.J. | | Deposit date: | 2013-10-21 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Molecular basis for unidirectional scaffold switching of human Plk4 in centriole biogenesis.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4LKM
 
 | | Crystal structure of Plk1 Polo-box domain in complex with PL-74 | | Descriptor: | GLYCEROL, PL-74, SULFATE ION, ... | | Authors: | Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2013-07-08 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the binding nature of pyrrolidine pocket-dependent interactions in the polo-box domain of polo-like kinase 1
Plos One, 8, 2013
|
|
4LKL
 
 | |
8YY8
 
 | | Fzd7 -Gs complex | | Descriptor: | Frizzled-7, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Chen, B, Xu, L, Han, G.W, Xu, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Cryo-EM structure of constitutively active human Frizzled 7 in complex with heterotrimeric G s .
Cell Res., 31, 2021
|
|
3KJF
 
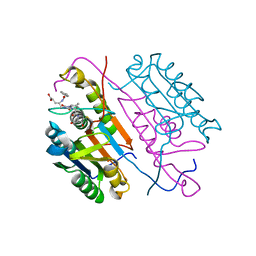 | | Caspase 3 Bound to a covalent inhibitor | | Descriptor: | (3S)-3-({[(5S,10aS)-2-{(2S)-4-carboxy-2-[(phenylacetyl)amino]butyl}-1,3-dioxo-2,3,5,7,8,9,10,10a-octahydro-1H-[1,2,4]triazolo[1,2-a]cinnolin-5-yl]carbonyl}amino)-4-oxopentanoic acid, Caspase-3 | | Authors: | Kamtekar, S, Watt, W, Finzel, B.C, Harris, M.S, Blinn, J, Wang, Z, Tomasselli, A.G. | | Deposit date: | 2009-11-03 | | Release date: | 2010-08-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural characterization of caspase-3 and caspase-8 inhibition by a novel class of irreversible inhibitors.
Biochim.Biophys.Acta, 1804, 2010
|
|
3KJN
 
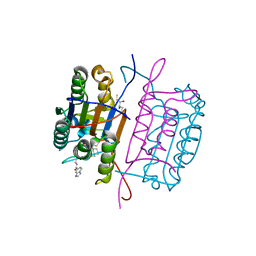 | | Caspase 8 bound to a covalent inhibitor | | Descriptor: | (3S)-3-({[(5S)-2-{2-[(1H-benzimidazol-5-ylcarbonyl)amino]ethyl}-7-(cyclohexylmethyl)-1,3-dioxo-2,3,5,8-tetrahydro-1H-[1,2,4]triazolo[1,2-a]pyridazin-5-yl]carbonyl}amino)-4-oxopentanoic acid, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Caspase-8 | | Authors: | Kamtekar, S, Watt, W, Finzel, B.C, Harris, M.S, Blinn, J, Wang, Z, Tomasselli, A.G. | | Deposit date: | 2009-11-03 | | Release date: | 2010-08-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and structural characterization of caspase-3 and caspase-8 inhibition by a novel class of irreversible inhibitors.
Biochim.Biophys.Acta, 1804, 2010
|
|
2CFO
 
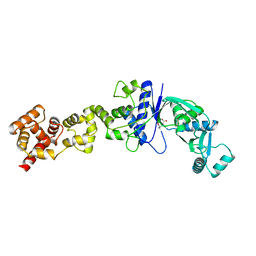 | | Non-Discriminating Glutamyl-tRNA Synthetase from Thermosynechococcus elongatus in Complex with Glu | | Descriptor: | GLUTAMIC ACID, GLUTAMYL-TRNA SYNTHETASE | | Authors: | Schulze, J.O, Nickel, D, Schubert, W.-D, Jahn, D, Heinz, D.W. | | Deposit date: | 2006-02-22 | | Release date: | 2006-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of a Non-Discriminating Glutamyl- tRNA Synthetase.
J.Mol.Biol., 361, 2006
|
|
6ENE
 
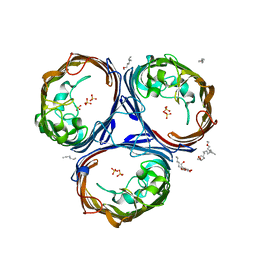 | | OmpF orthologue from Enterobacter cloacae (OmpE35) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Outer membrane protein (Porin), SULFATE ION, ... | | Authors: | van den Berg, B, Abellon-Ruiz, J, Basle, A. | | Deposit date: | 2017-10-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Getting Drugs into Gram-Negative Bacteria: Rational Rules for Permeation through General Porins.
Acs Infect Dis., 4, 2018
|
|
5O79
 
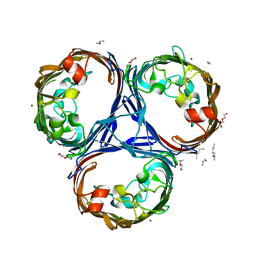 | | Klebsiella pneumoniae OmpK36 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MAGNESIUM ION, OmpK36 | | Authors: | van den berg, B, Pathania, M. | | Deposit date: | 2017-06-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Getting Drugs into Gram-Negative Bacteria: Rational Rules for Permeation through General Porins.
Acs Infect Dis., 4, 2018
|
|
1CGC
 
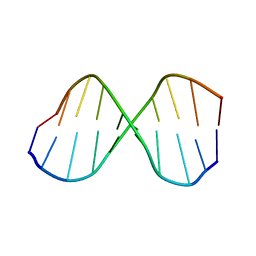 | |
3KJQ
 
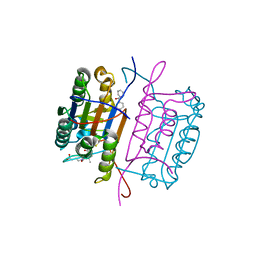 | | Caspase 8 with covalent inhibitor | | Descriptor: | (3S)-3-({[(5S,8R)-2-(3-carboxypropyl)-8-(2-{[(4-chlorophenyl)acetyl]amino}ethyl)-1,3-dioxo-2,3,5,8-tetrahydro-1H-[1,2,4]triazolo[1,2-a]pyridazin-5-yl]carbonyl}amino)-4-oxopentanoic acid, Caspase-8 | | Authors: | Kamtekar, S, Watt, W, Finzel, B.C, Harris, M.S, Blinn, J, Wang, Z, Tomasselli, A.G. | | Deposit date: | 2009-11-03 | | Release date: | 2010-08-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and structural characterization of caspase-3 and caspase-8 inhibition by a novel class of irreversible inhibitors.
Biochim.Biophys.Acta, 1804, 2010
|
|
3D6E
 
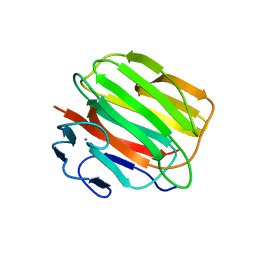 | | Crystal structure of the engineered 1,3-1,4-beta-glucanase protein from Bacillus licheniformis | | Descriptor: | Beta-glucanase, CALCIUM ION | | Authors: | Fita, I, Planas, A, Calisto, B.M, Addington, T. | | Deposit date: | 2008-05-19 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Re-engineering specificity in 1,3-1,4-beta-glucanase to accept branched xyloglucan substrates
Proteins, 79, 2011
|
|
5JW7
 
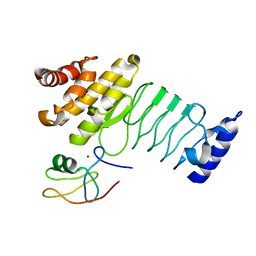 | | Crystal structure of SopA-Trim56 complex | | Descriptor: | E3 ubiquitin-protein ligase SopA, E3 ubiquitin-protein ligase TRIM56, ZINC ION | | Authors: | Bhogaraju, S, Dikic, I. | | Deposit date: | 2016-05-11 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.849 Å) | | Cite: | Structural basis for the recognition and degradation of host TRIM proteins by Salmonella effector SopA.
Nat Commun, 8, 2017
|
|
6ZLU
 
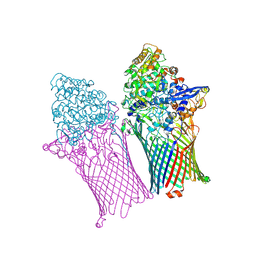 | |
6Z9A
 
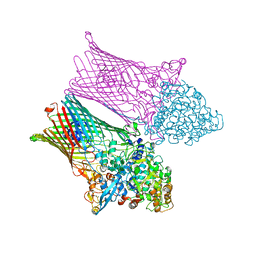 | | Fructo-oligosaccharide transporter BT 1762-63 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SusC homolog, ... | | Authors: | van den Berg, B. | | Deposit date: | 2020-06-03 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Insights into SusCD-mediated glycan import by a prominent gut symbiont.
Nat Commun, 12, 2021
|
|
6ZM1
 
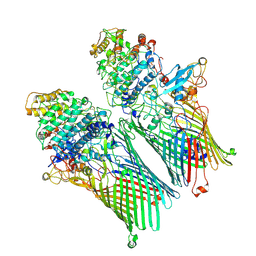 | |
6Z8I
 
 | | Fructo-oligosaccharide transporter BT 1762-63 | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, SusC homolog, ... | | Authors: | van den Berg, B. | | Deposit date: | 2020-06-02 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Insights into SusCD-mediated glycan import by a prominent gut symbiont.
Nat Commun, 12, 2021
|
|
6ZAZ
 
 | | Fructo-oligosaccharide transporter BT 1762-63 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | van den Berg, B. | | Deposit date: | 2020-06-06 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Insights into SusCD-mediated glycan import by a prominent gut symbiont.
Nat Commun, 12, 2021
|
|
6ZLT
 
 | |
6YTC
 
 | |
2ORB
 
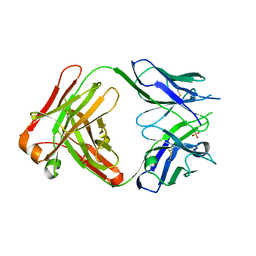 | | The structure of the anti-c-myc antibody 9E10 Fab fragment | | Descriptor: | Monoclonal anti-c-myc antibody 9E10, SULFATE ION | | Authors: | Krauss, N, Scheerer, P, Hoehne, W. | | Deposit date: | 2007-02-02 | | Release date: | 2008-02-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the anti-c-myc antibody 9E10 Fab fragment/epitope peptide complex reveals a novel binding mode dominated by the heavy chain hypervariable loops.
Proteins, 73, 2008
|
|
2OR9
 
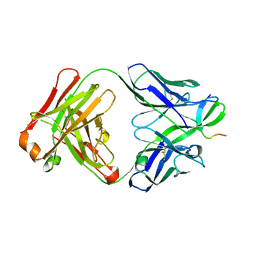 | |
1AJK
 
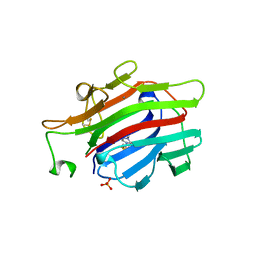 | | CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE CPA16M-84 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE, ... | | Authors: | Ay, J, Heinemann, U. | | Deposit date: | 1997-05-06 | | Release date: | 1998-05-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures and properties of de novo circularly permuted 1,3-1,4-beta-glucanases.
Proteins, 30, 1998
|
|
1AJO
 
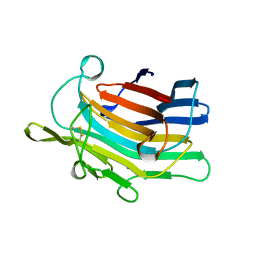 | | CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE CPA16M-127 | | Descriptor: | CALCIUM ION, CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE CPA16M-127 | | Authors: | Ay, J, Heinemann, U. | | Deposit date: | 1997-05-07 | | Release date: | 1998-05-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structures and properties of de novo circularly permuted 1,3-1,4-beta-glucanases.
Proteins, 30, 1998
|
|
3HAT
 
 | | ACTIVE SITE MIMETIC INHIBITION OF THROMBIN | | Descriptor: | FPAM (FIBRINOPEPTIDE A MIMIC), Hirudin variant-2, Thrombin heavy chain, ... | | Authors: | Tulinsky, A, Mathews, I.I. | | Deposit date: | 1994-10-16 | | Release date: | 1995-02-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Active-site mimetic inhibition of thrombin.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
