4WJU
 
 | | Crystal structure of Rsa4 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Ribosome assembly protein 4 | | Authors: | Holdermann, I, Bassler, J, Hurt, E, Sinning, I. | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
1C50
 
 | | IDENTIFICATION AND STRUCTURAL CHARACTERIZATION OF A NOVEL ALLOSTERIC BINDING SITE OF GLYCOGEN PHOSPHORYLASE B | | Descriptor: | 5-CHLORO-1H-INDOLE-2-CARBOXYLIC ACID [1-(4-FLUOROBENZYL)-2-(4-HYDROXYPIPERIDIN-1YL)-2-OXOETHYL]AMIDE, PROTEIN (GLYCOGEN PHOSPHORYLASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Oikonomakos, N.G, Skamnaki, V.T, Tsitsanou, K.E, Gavalas, N.G, Johnson, L.N. | | Deposit date: | 1999-12-15 | | Release date: | 1999-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A new allosteric site in glycogen phosphorylase b as a target for drug interactions.
Structure Fold.Des., 8, 2000
|
|
1C8K
 
 | | FLAVOPIRIDOL INHIBITS GLYCOGEN PHOSPHORYLASE BY BINDING AT THE INHIBITOR SITE | | Descriptor: | 2-(2-CHLORO-PHENYL)-5,7-DIHYDROXY-8-(3-HYDROXY-1-METHYL-PIPERIDIN-4-YL)-4H-BENZOPYRAN-4-ONE, PROTEIN (GLYCOGEN PHOSPHORYLASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Oikonomakos, N.G, Zographos, S.E, Skamnaki, V.T, Tsitsanou, K.E, Johnson, L.N. | | Deposit date: | 2000-05-11 | | Release date: | 2000-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Flavopiridol inhibits glycogen phosphorylase by binding at the inhibitor site.
J.Biol.Chem., 275, 2000
|
|
1C7D
 
 | | DEOXY RHB1.2 (RECOMBINANT HEMOGLOBIN) | | Descriptor: | PROTEIN (DEOXYHEMOGLOBIN (ALPHA CHAIN)), PROTEIN (DEOXYHEMOGLOBIN (BETA CHAIN)), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Brucker, E.A. | | Deposit date: | 2000-02-09 | | Release date: | 2000-06-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Genetically crosslinked hemoglobin: a structural study.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
6Q7A
 
 | |
6Q6O
 
 | |
1C7C
 
 | | DEOXY RHB1.1 (RECOMBINANT HEMOGLOBIN) | | Descriptor: | PROTEIN (DEOXYHEMOGLOBIN (ALPHA CHAIN)), PROTEIN (DEOXYHEMOGLOBIN (BETA CHAIN)), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Brucker, E.A. | | Deposit date: | 2000-02-09 | | Release date: | 2000-06-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Genetically crosslinked hemoglobin: a structural study.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1C7B
 
 | | DEOXY RHB1.0 (RECOMBINANT HEMOGLOBIN) | | Descriptor: | PROTEIN (DEOXYHEMOGLOBIN (ALPHA CHAIN)), PROTEIN (DEOXYHEMOGLOBIN (BETA CHAIN)), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Brucker, E.A. | | Deposit date: | 2000-02-09 | | Release date: | 2000-06-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Genetically crosslinked hemoglobin: a structural study.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
6Q7H
 
 | |
6Q6M
 
 | |
1CA5
 
 | | INTERCALATION SITE OF HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SSO7D/SAC7D BOUND TO DNA | | Descriptor: | 5'-D(*GP*TP*GP*AP*TP*CP*AP*C)-3', CHROMOSOMAL PROTEIN SAC7D | | Authors: | Su, S, Gao, Y.-G, Robinson, H, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 1999-02-23 | | Release date: | 2000-02-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the chromosomal proteins Sso7d/Sac7d bound to DNA containing T-G mismatched base-pairs
J.Mol.Biol., 303, 2000
|
|
4BI6
 
 | | CRYSTAL STRUCTURE OF A TRIPLE MUTANT (A198V, C202A AND C222N) OF TRIOSEPHOSPHATE ISOMERASE FROM GIARDIA LAMBLIA. COMPLEXED WITH 2- PHOSPHOGLYCOLIC ACID | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Torres-Larios, A, Enriquez-Flores, S, Reyes-Vivas, H, Oria-Hernandez, J, Hernandez-Alcantara, G. | | Deposit date: | 2013-04-09 | | Release date: | 2013-05-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Functional Perturbation of Giardia Lamblia Triosephosphate Isomerase by Modification of a Non-Catalytic, Non-Conserved Region.
Plos One, 8, 2013
|
|
4BI7
 
 | | CRYSTAL STRUCTURE OF A MUTANT (C202A) OF TRIOSEPHOSPHATE ISOMERASE FROM GIARDIA LAMBLIA COMPLEXED WITH 2-PHOSPHOGLYCOLIC ACID | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, SULFATE ION, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Torres-Larios, A, Enriquez-Flores, S, Reyes-Vivas, H, Oria-Hernandez, J, Hernandez-Alcantara, G. | | Deposit date: | 2013-04-09 | | Release date: | 2013-05-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Perturbation of Giardia Lamblia Triosephosphate Isomerase by Modification of a Non-Catalytic, Non-Conserved Region.
Plos One, 8, 2013
|
|
4BI5
 
 | | CRYSTAL STRUCTURE OF A DOUBLE MUTANT (C202A AND C222D) OF TRIOSEPHOSPHATE ISOMERASE FROM GIARDIA LAMBLIA. | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Torres-Larios, A, Enriquez-Flores, S, Reyes-Vivas, H, Oria-Hernandez, J, Hernandez-Alcantara, G. | | Deposit date: | 2013-04-09 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Perturbation of Giardia Lamblia Triosephosphate Isomerase by Modification of a Non-Catalytic, Non-Conserved Region.
Plos One, 8, 2013
|
|
7B17
 
 | | SARS-CoV-spike RBD bound to two neutralising nanobodies. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SARS-CoV-2 neutralizing biparatopic nanobody VE,nanobody E from Lama glama,SARS-CoV-2 neutralizing biparatopic nanobody VE,nanobody E from Lama glama, Spike protein S1 | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2020-11-23 | | Release date: | 2021-02-10 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape
Science, 371, 2021
|
|
7OAJ
 
 | | Crystal structure of pseudokinase CASK in complex with compound 7 | | Descriptor: | 1,2-ETHANEDIOL, 4-(cyclopentylamino)-2-[(3,4-dichlorophenyl)methylamino]-N-[3-(2-oxidanylidenepyrrolidin-1-yl)propyl]pyrimidine-5-carboxamide, Peripheral plasma membrane protein CASK | | Authors: | Chaikuad, A, Russ, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Design and Development of a Chemical Probe for Pseudokinase Ca 2+ /calmodulin-Dependent Ser/Thr Kinase.
J.Med.Chem., 64, 2021
|
|
7OAI
 
 | | Crystal structure of pseudokinase CASK in complex with PFE-PKIS12 | | Descriptor: | 1,2-ETHANEDIOL, 4-(Cyclopentylamino)-2-[(2,5-dichlorophenyl)methylamino]-N-[3-(2-oxo-1,3-oxazolidin-3-yl)propyl]pyrimidine-5-carboxamide, Peripheral plasma membrane protein CASK | | Authors: | Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Development of a Chemical Probe for Pseudokinase Ca 2+ /calmodulin-Dependent Ser/Thr Kinase.
J.Med.Chem., 64, 2021
|
|
7OAL
 
 | | Crystal structure of pseudokinase CASK in complex with compound 25 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[2,5-bis(bromanyl)-4-methyl-phenyl]methylamino]-4-(cyclohexylamino)-N-[3-(2-oxidanylidene-1,3-oxazolidin-3-yl)propyl]pyrimidine-5-carboxamide, Peripheral plasma membrane protein CASK | | Authors: | Chaikuad, A, Russ, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Design and Development of a Chemical Probe for Pseudokinase Ca 2+ /calmodulin-Dependent Ser/Thr Kinase.
J.Med.Chem., 64, 2021
|
|
7OAK
 
 | | Crystal structure of pseudokinase CASK in complex with compound 26 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[2,5-bis(bromanyl)-4-methyl-phenyl]methylamino]-4-(cyclopentylamino)-N-[3-(2-oxidanylidene-1,3-oxazolidin-3-yl)propyl]pyrimidine-5-carboxamide, Peripheral plasma membrane protein CASK | | Authors: | Chaikuad, A, Russ, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Design and Development of a Chemical Probe for Pseudokinase Ca 2+ /calmodulin-Dependent Ser/Thr Kinase.
J.Med.Chem., 64, 2021
|
|
7OAM
 
 | | Kinase domain of MERTK in complex with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[2,5-bis(fluoranyl)phenyl]methylamino]-4-(cyclopentylamino)-N-[3-(2-oxidanylidenepyrrolidin-1-yl)propyl]pyrimidine-5-carboxamide, Tyrosine-protein kinase Mer | | Authors: | Schroeder, M, Russ, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Design and Development of a Chemical Probe for Pseudokinase Ca 2+ /calmodulin-Dependent Ser/Thr Kinase.
J.Med.Chem., 64, 2021
|
|
7B14
 
 | | Nanobody E bound to Spike-RBD in a localized reconstruction | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody against SARS-CoV-2, Spike protein S1 | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2020-11-23 | | Release date: | 2021-04-28 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape
Science, 371, 2021
|
|
7B18
 
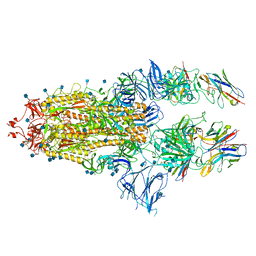 | | SARS-CoV-spike bound to two neutralising nanobodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody against SARS-CoV-2 VHH E, ... | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
4WJS
 
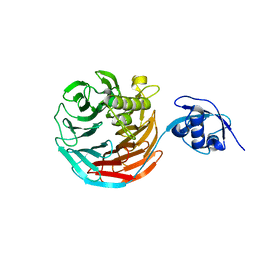 | |
8S7J
 
 | | Coxsackievirus A9 bound with compound 20 (CL300) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Plavec, Z, Butcher, S.J, Mitchell, C, Buckner, C. | | Deposit date: | 2024-03-01 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | SAR Analysis of Novel Coxsackie virus A9 Capsid Binders.
J.Med.Chem., 2024
|
|
1SIQ
 
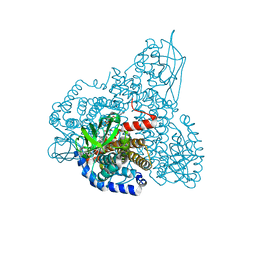 | | The Crystal Structure and Mechanism of Human Glutaryl-CoA Dehydrogenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase | | Authors: | Wang, M, Fu, Z, Paschke, R, Goodman, S, Frerman, F.E, Kim, J.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Human Glutaryl-CoA Dehydrogenase with and without an Alternate Substrate: Structural Bases of Dehydrogenation and Decarboxylation Reactions
Biochemistry, 43, 2004
|
|
