2HWR
 
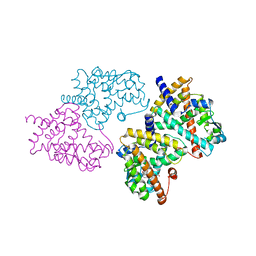 | | Structural basis for the structure-activity relationships of Peroxisome Proliferator-Activated Receptor agonists | | Descriptor: | 2-[(1-{3-[(6-BENZOYL-1-PROPYL-2-NAPHTHYL)OXY]PROPYL}-1H-INDOL-4-YL)OXY]-2-METHYLPROPANOIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Peng, Y.H, Lu, I.L, Mahindroo, N, Lin, C.H, Hsieh, H.P, Wu, S.Y. | | Deposit date: | 2006-08-01 | | Release date: | 2007-08-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for the structure-activity relationships of peroxisome proliferator-activated receptor agonists
J.Med.Chem., 49, 2006
|
|
2HWQ
 
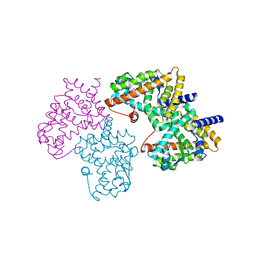 | | Structural basis for the structure-activity relationships of Peroxisome Proliferator-Activated Receptor agonists | | Descriptor: | Peroxisome proliferator-activated receptor gamma, [(1-{3-[(6-BENZOYL-1-PROPYL-2-NAPHTHYL)OXY]PROPYL}-1H-INDOL-5-YL)OXY]ACETIC ACID | | Authors: | Peng, Y.H, Lu, I.L, Mahindroo, N, Lin, C.H, Hsieh, H.P, Wu, S.Y. | | Deposit date: | 2006-08-01 | | Release date: | 2007-08-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis for the structure-activity relationships of peroxisome proliferator-activated receptor agonists
J.Med.Chem., 49, 2006
|
|
2IHK
 
 | |
2IHJ
 
 | |
2IIQ
 
 | |
2ILV
 
 | |
2IHZ
 
 | |
2II6
 
 | |
6B76
 
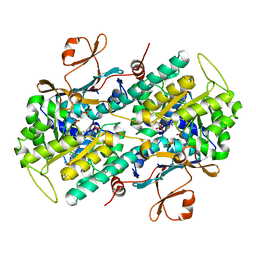 | | Crystal Structure of human NAMPT in complex with NVP-LVR596 | | Descriptor: | (1S,2S)-N-{4-[(1S)-1-(propanoylamino)ethyl]phenyl}-2-(pyridin-3-yl)cyclopropane-1-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Weihofen, W.A, Thigale, S. | | Deposit date: | 2017-10-03 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Identification and structure based design of cellularly active cyclo-propyl carboxamide Nicotinamide phosphoribosyltransferase (NAMPT) inhibitors
To Be Published
|
|
7WKH
 
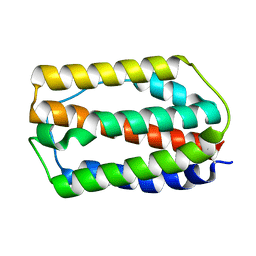 | | the grass carp IFNa | | Descriptor: | Interferon | | Authors: | Zou, J, WANG, J, Wang, Z. | | Deposit date: | 2022-01-09 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural and Functional Analyses of Type I IFNa Shed Light Into Its Interaction With Multiple Receptors in Fish.
Front Immunol, 13, 2022
|
|
6JBZ
 
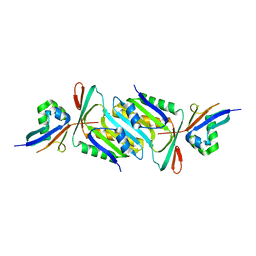 | |
8WRH
 
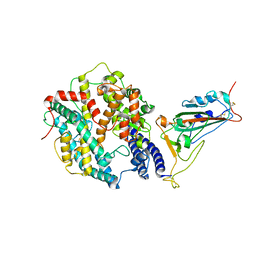 | | SARS-CoV-2 XBB.1.5.70 in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S2' | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-14 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WRL
 
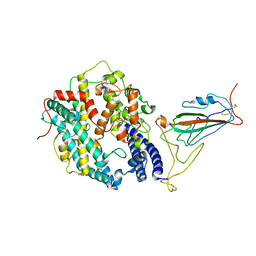 | | XBB.1.5 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-15 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
6JC0
 
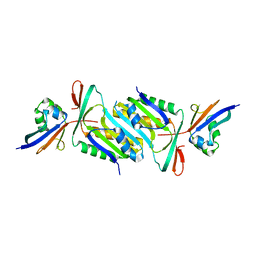 | |
8WRM
 
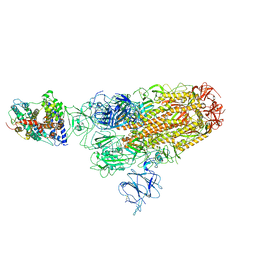 | | XBB.1.5 spike protein in complex with ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-15 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WRO
 
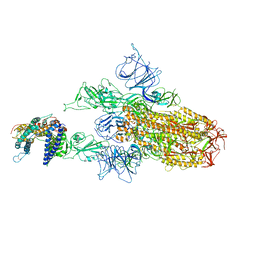 | | XBB.1.5.10 spike protein in complex with ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike glycoprotein,Spike glycoprotein,Spike glycoprotein,Fusion protein | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-15 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WTD
 
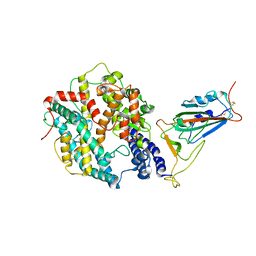 | | XBB.1.5.10 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S2' | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WTJ
 
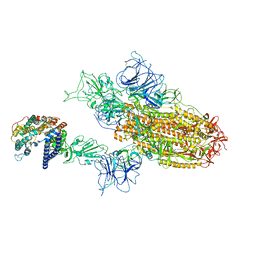 | | XBB.1.5.70 spike protein in complex with ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.64 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WUY
 
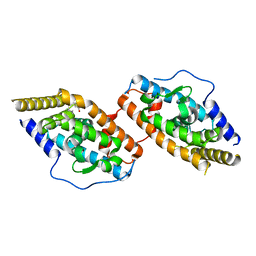 | | Crystal Structure of TR3 LBD in complex with para-positioned 3,4,5-trisubstituted benzene derivatives | | Descriptor: | Nuclear receptor subfamily 4immunitygroup A member 1, ~{N}-methyl-~{N}-octyl-3,4,5-tris(oxidanyl)benzamide | | Authors: | Hong, W.B, Chen, X.Q, Lin, T.W. | | Deposit date: | 2023-10-21 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design and synthesis of anti-fibrotic compounds derived from para-positioned 3,4,5-trisubstituted benzene.
Bioorg.Chem., 144, 2024
|
|
3HSZ
 
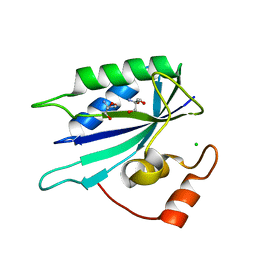 | | Crystal structure of E. coli HPPK(F123A) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-06-11 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Pterin-binding site mutation Y53A, N55A or F123A and activity of E. coli HPPK
To be Published
|
|
5E66
 
 | | The complex structure of Hemagglutinin-esterase-fusion mutant protein from the influenza D virus with receptor analog 9-N-Ac-Sia | | Descriptor: | (6R)-5-acetamido-6-[(1S,2S)-3-acetamido-1,2-dihydroxypropyl]-3,5-dideoxy-beta-L-threo-hex-2-ulopyranosonic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-10-09 | | Release date: | 2016-03-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism
PLoS Pathog., 12, 2016
|
|
5DRZ
 
 | | Crystal structure of anti-HIV-1 antibody F240 Fab in complex with gp41 peptide | | Descriptor: | Envelope glycoprotein gp160, HIV Antibody F240 Heavy Chain, HIV Antibody F240 Light Chain, ... | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2015-09-16 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Molecular basis for epitope recognition by non-neutralizing anti-gp41 antibody F240.
Sci Rep, 6, 2016
|
|
5E62
 
 | | HEF-mut with Tr323 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-azidoethyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, ... | | Authors: | Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-10-09 | | Release date: | 2016-03-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism
PLoS Pathog., 12, 2016
|
|
5E65
 
 | | The complex structure of Hemagglutinin-esterase-fusion mutant protein from the influenza D virus with receptor analog 9-O-Ac-3'SLN (Tr322) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-10-09 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism
PLoS Pathog., 12, 2016
|
|
5E64
 
 | | Hemagglutinin-esterase-fusion protein structure of influenza D virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, ... | | Authors: | Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-10-09 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism
PLoS Pathog., 12, 2016
|
|
