2F3A
 
 | |
2FBU
 
 | | Solution structure of the N-terminal fragment of human LL-37 | | Descriptor: | Antibacterial protein FALL-39, core peptide | | Authors: | Wang, G, Li, X. | | Deposit date: | 2005-12-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Human LL-37 Fragments and NMR-Based Identification of a Minimal Membrane-Targeting Antimicrobial and Anticancer Region
J.Am.Chem.Soc., 128, 2006
|
|
2FBS
 
 | |
7LIB
 
 | |
7N4X
 
 | | Structure of human NPC1L1 mutant-W347R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Li, X, Long, T. | | Deposit date: | 2021-06-04 | | Release date: | 2021-09-01 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structures of dimeric human NPC1L1 provide insight into mechanisms for cholesterol absorption.
Sci Adv, 7, 2021
|
|
7N4U
 
 | | Structure of human NPC1L1 | | Descriptor: | (2R)-2,5,7,8-TETRAMETHYL-2-[(4R,8R)-4,8,12-TRIMETHYLTRIDECYL]CHROMAN-6-OL, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, X, Long, T. | | Deposit date: | 2021-06-04 | | Release date: | 2021-09-01 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structures of dimeric human NPC1L1 provide insight into mechanisms for cholesterol absorption.
Sci Adv, 7, 2021
|
|
7N4V
 
 | | Structure of cholesterol-bound human NPC1L1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, X, Long, T. | | Deposit date: | 2021-06-04 | | Release date: | 2021-09-01 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structures of dimeric human NPC1L1 provide insight into mechanisms for cholesterol absorption.
Sci Adv, 7, 2021
|
|
5EE9
 
 | | Complex structure of OSYCHF1 with GMP-PNP | | Descriptor: | GLYCEROL, MAGNESIUM ION, Obg-like ATPase 1, ... | | Authors: | Li, X, Chen, Z. | | Deposit date: | 2015-10-22 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | ATP binding by the P-loop NTPase OsYchF1 (an unconventional G protein) contributes to biotic but not abiotic stress responses
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7XST
 
 | |
5EE3
 
 | | COMPLEX STRUCTURE OF OSYCHF1 WITH AMP-PNP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Li, X, Chen, Z. | | Deposit date: | 2015-10-22 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | ATP binding by the P-loop NTPase OsYchF1 (an unconventional G protein) contributes to biotic but not abiotic stress responses
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7XMX
 
 | | Cryo-EM structure of SARS-CoV-2 spike glycoprotein in complex with three F61 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F61 heavy chain, F61 light chain, ... | | Authors: | Wang, X, Li, X. | | Deposit date: | 2022-04-27 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis of a two-antibody cocktail exhibiting highly potent and broadly neutralizing activities against SARS-CoV-2 variants including diverse Omicron sublineages.
Cell Discov, 8, 2022
|
|
7XMZ
 
 | | Cryo-EM structure of SARS-CoV-2 spike glycoprotein in complex with three D2 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D2 heavy chain, D2 light chain, ... | | Authors: | Wang, X, Li, X. | | Deposit date: | 2022-04-27 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis of a two-antibody cocktail exhibiting highly potent and broadly neutralizing activities against SARS-CoV-2 variants including diverse Omicron sublineages.
Cell Discov, 8, 2022
|
|
5EE1
 
 | | Crystal structure of OsYchF1 at pH 7.85 | | Descriptor: | Obg-like ATPase 1 | | Authors: | Li, X, Chen, Z. | | Deposit date: | 2015-10-22 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | ATP binding by the P-loop NTPase OsYchF1 (an unconventional G protein) contributes to biotic but not abiotic stress responses
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EE0
 
 | | Crystal structure of OsYchF1 at pH 6.5 | | Descriptor: | Obg-like ATPase 1 | | Authors: | Li, X, Chen, Z. | | Deposit date: | 2015-10-22 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | ATP binding by the P-loop NTPase OsYchF1 (an unconventional G protein) contributes to biotic but not abiotic stress responses
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4TVQ
 
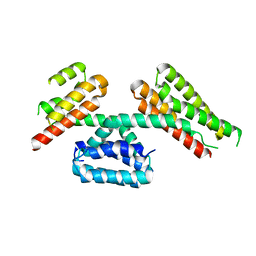 | | CCM3 in complex with CCM2 LD-like motif | | Descriptor: | Cerebral cavernous malformations 2 protein, Cerebral cavernous malformations 3 protein | | Authors: | Li, X, Zhang, R, Fisher, O.S, Boggon, T.J. | | Deposit date: | 2014-06-27 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CCM2-CCM3 interaction stabilizes their protein expression and permits endothelial network formation.
J.Cell Biol., 208, 2015
|
|
6N7R
 
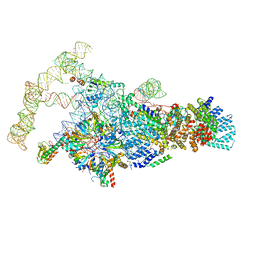 | | Saccharomyces cerevisiae spliceosomal E complex (ACT1) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, ACT1 pre-mRNA, Pre-mRNA-processing factor 39, ... | | Authors: | Liu, S, Li, X, Zhou, Z.H, Zhao, R. | | Deposit date: | 2018-11-28 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A unified mechanism for intron and exon definition and back-splicing.
Nature, 573, 2019
|
|
4YMQ
 
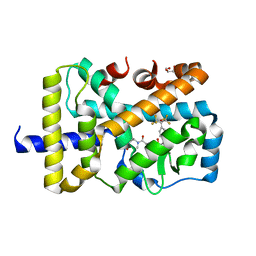 | | X-ray co-structure of nuclear receptor ROR-GAMMAT + SRC2 peptide with a benzothiadiazole dioxide inverse agonist | | Descriptor: | 4-{3-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)benzyl]-2,2-dioxido-2,1,3-benzothiadiazol-1(3H)-yl}-N-[(2R)-4-hydroxybutan-2-yl]-N-methylbutanamide, GLYCEROL, Nuclear receptor ROR-gamma, ... | | Authors: | li, X. | | Deposit date: | 2015-03-07 | | Release date: | 2015-04-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 1,3-dihydro-2,1,3-benzothiadiazole 2,2-dioxide analogs as new RORC modulators.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
3NDM
 
 | |
5YGI
 
 | |
2LEF
 
 | | LEF1 HMG DOMAIN (FROM MOUSE), COMPLEXED WITH DNA (15BP), NMR, 12 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*AP*CP*CP*CP*TP*TP*TP*GP*AP*AP*GP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*CP*TP*TP*CP*AP*AP*AP*GP*GP*GP*TP*G)-3'), PROTEIN (LYMPHOID ENHANCER-BINDING FACTOR) | | Authors: | Li, X, Love, J.J, Case, D.A, Wright, P.E. | | Deposit date: | 1998-10-13 | | Release date: | 1998-10-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis for DNA bending by the architectural transcription factor LEF-1.
Nature, 376, 1995
|
|
7WJQ
 
 | | Crystal structure of GSDMB in complex with Ipah7.8 | | Descriptor: | Isoform 2 of Gasdermin-B, Probable E3 ubiquitin-protein ligase ipaH7.8 | | Authors: | Li, X, Zhang, H, Yin, H. | | Deposit date: | 2022-01-07 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into the GSDMB-mediated cellular lysis and its targeting by IpaH7.8
Nat Commun, 14, 2023
|
|
2FLU
 
 | | Crystal Structure of the Kelch-Neh2 Complex | | Descriptor: | Kelch-like ECH-associated protein 1, Nrf2 | | Authors: | Li, X, Lo, J, Beamer, L, Hannink, M. | | Deposit date: | 2006-01-06 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling.
Embo J., 25, 2006
|
|
3UR1
 
 | |
3NCZ
 
 | |
7Y9I
 
 | | Complex structure of AtYchF1 with ppGpp | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, MAGNESIUM ION, Obg-like ATPase 1 | | Authors: | Li, X, Chen, Z. | | Deposit date: | 2022-06-24 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Co-crystalization reveals the interaction between AtYchF1 and ppGpp.
Front Mol Biosci, 9, 2022
|
|
