7MC4
 
 | |
2C74
 
 | | 14-3-3 Protein Eta (Human) Complexed to Peptide | | Descriptor: | 14-3-3 PROTEIN ETA, CITRIC ACID, CONSENSUS PEPTIDE MODE 1 FOR 14-3-3 PROTEINS | | Authors: | Elkins, J.M, Yang, X, Smee, C.E.A, Johansson, C, Sundstrom, M, Edwards, A, Weigelt, J, Arrowsmith, C, Doyle, D.A. | | Deposit date: | 2005-11-17 | | Release date: | 2005-12-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for protein-protein interactions in the 14-3-3 protein family.
Proc. Natl. Acad. Sci. U.S.A., 103, 2006
|
|
2C23
 
 | | 14-3-3 Protein Beta (Human) in complex with exoenzyme S peptide | | Descriptor: | 14-3-3 BETA/ALPHA, EXOENZYME S PEPTIDE | | Authors: | Elkins, J.M, Schoch, G.A, Yang, X, Sundstrom, M, Arrowsmith, C, Edwards, A, Doyle, D.A. | | Deposit date: | 2005-09-26 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Basis for Protein-Protein Interactions in the 14-3-3 Protein Family.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
1VR4
 
 | | Crystal Structure of MCSG TArget APC22750 from Bacillus cereus | | Descriptor: | hypothetical protein APC22750 | | Authors: | Yang, X, Brunzelle, J.S, McNamara, L.K, Minasov, G, Shuvalova, L, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-01-31 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of MCSG TArget APC22750 from Bacillus cereus
To be Published
|
|
4NBM
 
 | | Crystal structure of UVB photoreceptor UVR8 and light-induced structural changes at 180K | | Descriptor: | MAGNESIUM ION, Ultraviolet-B receptor UVR8 | | Authors: | Yang, X, Zeng, X, Zhao, K.-H, Ren, Z. | | Deposit date: | 2013-10-23 | | Release date: | 2016-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Dynamic Crystallography Reveals Early Signalling Events in Ultraviolet Photoreceptor UVR8.
Nat Plants, 1, 2015
|
|
4NAA
 
 | | Crystal structure of UVB photoreceptor UVR8 from Arabidopsis thaliana and UV-induced structural changes at 120K | | Descriptor: | MAGNESIUM ION, Ultraviolet-B receptor UVR8 | | Authors: | Yang, X, Zeng, X, Ren, Z, Zhao, K.H. | | Deposit date: | 2013-10-22 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Dynamic Crystallography Reveals Early Signalling Events in Ultraviolet Photoreceptor UVR8.
Nat Plants, 1, 2015
|
|
4NC4
 
 | | Crystal structure of photoreceptor AtUVR8 mutant W285F and light-induced structural changes at 120K | | Descriptor: | MAGNESIUM ION, Ultraviolet-B receptor UVR8 | | Authors: | Yang, X, Zeng, X, Zhao, K.-H, Ren, Z. | | Deposit date: | 2013-10-23 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dynamic Crystallography Reveals Early Signalling Events in Ultraviolet Photoreceptor UVR8.
Nat Plants, 1, 2015
|
|
2BTP
 
 | | 14-3-3 Protein Theta (Human) Complexed to Peptide | | Descriptor: | 14-3-3 PROTEIN TAU, CONSENSUS PEPTIDE FOR 14-3-3 PROTEINS | | Authors: | Elkins, J.M, Johansson, A.C.E, Smee, C, Yang, X, Sundstrom, M, Edwards, A, Arrowsmith, C, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-05 | | Release date: | 2005-06-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Protein-Protein Interactions in the 14-3-3 Protein Family.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
5XGH
 
 | | Crystal structure of PI3K complex with an inhibitor | | Descriptor: | 3-[(4-fluorophenyl)methylamino]-5-(4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)phenol, GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Song, K, Yang, X, Zhao, Y, Jian, Z. | | Deposit date: | 2017-04-13 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | New Insights into PI3K Inhibitor Design using X-ray Structures of PI3K alpha Complexed with a Potent Lead Compound.
Sci Rep, 7, 2017
|
|
2ODB
 
 | | The crystal structure of human cdc42 in complex with the CRIB domain of human p21-activated kinase 6 (PAK6) | | Descriptor: | CHLORIDE ION, Human Cell Division Cycle 42 (CDC42), MAGNESIUM ION, ... | | Authors: | Ugochukwu, E, Yang, X, Elkins, J, Soundararajan, M, Pike, A.C.W, Eswaran, J, Burgess, N, Debreczeni, J.E, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, Gileadi, O, von Delft, F, Knapp, S, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-22 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of human cdc42 in complex with the CRIB domain of human p21-activated kinase 6 (PAK6)
To be Published
|
|
2IC5
 
 | | Crystal structure of human RAC3 grown in the presence of Gpp(NH)p. | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ugochukwu, E, Yang, X, Zao, Y, Elkins, J, Gileadi, C, Burgess, N, Colebrook, S, Gileadi, O, Fedorov, O, Bunkoczi, G, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, von Delft, F, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-09-12 | | Release date: | 2006-10-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human RAC3 grown in the presence of Gpp(NH)p.
To be Published
|
|
8F71
 
 | |
7C2M
 
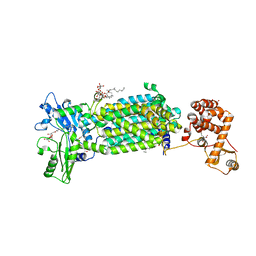 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with NITD-349 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Chimera of drug exporters of the RND superfamily-like protein and Endolysin, N-(4,4-dimethylcyclohexyl)-4,6-bis(fluoranyl)-1H-indole-2-carboxamide, ... | | Authors: | Zhang, B, Yang, X, Hu, T, Rao, Z. | | Deposit date: | 2020-05-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for the Inhibition of Mycobacterial MmpL3 by NITD-349 and SPIRO.
J.Mol.Biol., 432, 2020
|
|
7C2N
 
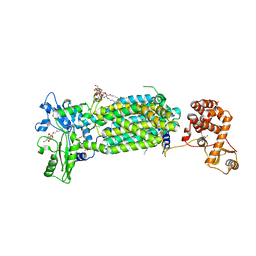 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with SPIRO | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 1'-(2,3-dihydro-1,4-benzodioxin-6-ylmethyl)spiro[6,7-dihydrothieno[3,2-c]pyran-4,4'-piperidine], Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Yang, X, Hu, T, Rao, Z. | | Deposit date: | 2020-05-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural Basis for the Inhibition of Mycobacterial MmpL3 by NITD-349 and SPIRO.
J.Mol.Biol., 432, 2020
|
|
7XRW
 
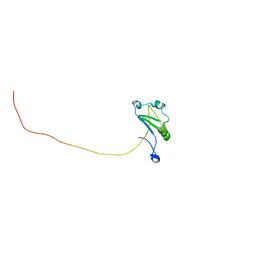 | | Solution structure of T. brucei RAP1 | | Descriptor: | Repressor activator protein 1 | | Authors: | Yang, X, Pan, X.H, Ji, Z.Y, Wong, K.B, Zhang, M.J, Zhao, Y.X. | | Deposit date: | 2022-05-12 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The RRM-mediated RNA binding activity in T. brucei RAP1 is essential for VSG monoallelic expression.
Nat Commun, 14, 2023
|
|
8YLN
 
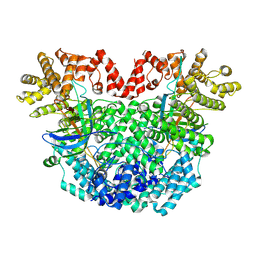 | | The structure of DSR2-Tail tube complex | | Descriptor: | Bacillus phage SPR Tube protein, SIR2-like domain-containing protein | | Authors: | Zheng, J, Yang, X. | | Deposit date: | 2024-03-06 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural insights into autoinhibition and activation of defense-associated sirtuin protein.
Int.J.Biol.Macromol., 277, 2024
|
|
8Z18
 
 | |
8YLT
 
 | | The structure of DSR2 and NAD+ complex | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SIR2-like domain-containing protein | | Authors: | Zheng, J, Yang, X. | | Deposit date: | 2024-03-06 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural insights into autoinhibition and activation of defense-associated sirtuin protein.
Int.J.Biol.Macromol., 277, 2024
|
|
8YKF
 
 | | The DSR2-DSAD1 complex with DSAD1 on the opposite sides | | Descriptor: | DSAD1, SIR2-like domain-containing protein | | Authors: | Zheng, J, Yang, X. | | Deposit date: | 2024-03-05 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural insights into autoinhibition and activation of defense-associated sirtuin protein.
Int.J.Biol.Macromol., 277, 2024
|
|
8ZTR
 
 | |
8STB
 
 | | The structure of abxF, an enzyme catalyzing the formation of the chiral spiroketal of an anthrabenzoxocinone antibiotic, (-)-ABX | | Descriptor: | CHLORIDE ION, GLYCEROL, Glyoxalase, ... | | Authors: | Luo, Z, Jia, X, Yan, X, Qu, X, Kobe, B. | | Deposit date: | 2023-05-09 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The crystal structure of abxF, an enzyme catalyzing the formation of the chiral spiroketal of an anthrabenzoxocinone antibiotic, (-)-ABX.
To Be Published
|
|
2X7I
 
 | | Crystal structure of mevalonate kinase from methicillin-resistant Staphylococcus aureus MRSA252 | | Descriptor: | CHLORIDE ION, CITRIC ACID, MEVALONATE KINASE | | Authors: | Oke, M, Yan, X, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-27 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
6DXK
 
 | | Glucocorticoid Receptor in complex with Compound 11 | | Descriptor: | (8S,11R,13S,14S,17S)-11-[4-(dimethylamino)phenyl]-17-(3,3-dimethylbut-1-yn-1-yl)-17-hydroxy-13-methyl-1,2,6,7,8,11,12,13,14,15,16,17-dodecahydro-3H-cyclopenta[a]phenanthren-3-one (non-preferred name), Glucocorticoid receptor | | Authors: | Rew, Y, Du, X, Eksterowicz, J, Zhou, H, Jahchan, N, Zhu, L, Yan, X, Kawai, H, McGee, L.R, Medina, J.C, Huang, T, Chen, C, Zavorotinskaya, T, Sutimantanapi, D, Waszczuk, J, Jackson, E, Huang, E, Ye, Q, Fantin, V.R, Daqing, S. | | Deposit date: | 2018-06-29 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Discovery of a Potent and Selective Steroidal Glucocorticoid Receptor Antagonist (ORIC-101).
J. Med. Chem., 61, 2018
|
|
8EPY
 
 | | The solution structure of abxF in complex with its product (-)-ABX, an enzyme catalyzing the formation of the chiral spiroketal of an anthrabenzoxocinone antibiotic, (-)-ABX | | Descriptor: | (6R,16R)-3,11,13,15-tetrahydroxy-1,6,9,9-tetramethyl-6,7,9,16-tetrahydro-14H-6,16-epoxyanthra[2,3-e]benzo[b]oxocin-14-one, Glyoxalase | | Authors: | Jia, X, Yan, X, Qu, X, Mobli, M. | | Deposit date: | 2022-10-06 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of abxF, an enzyme catalyzing the formation of chiral spiroketal of an antibiotics, (-)-ABX.
To Be Published
|
|
6CLW
 
 | | Crystal structure of TnmH | | Descriptor: | O-methyltransferase | | Authors: | Chang, C.Y, Annaval, T, Adhikari, A, Yan, X, Shen, B. | | Deposit date: | 2018-03-02 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Characterization of TnmH as anO-Methyltransferase Revealing Insights into Tiancimycin Biosynthesis and Enabling a Biocatalytic Strategy To Prepare Antibody-Tiancimycin Conjugates.
J.Med.Chem., 63, 2020
|
|
