2WD3
 
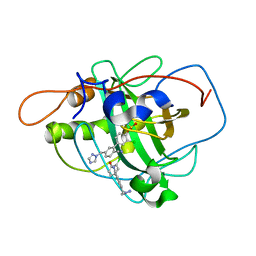 | | Highly Potent First Examples of Dual Aromatase-Steroid Sulfatase Inhibitors based on a Biphenyl Template | | Descriptor: | 3-CHLORO-2'-CYANO-5'-(1H-1,2,4-TRIAZOL-1-YLMETHYL)BIPHENYL-4-YL SULFAMATE, CARBONIC ANHYDRASE 2, ZINC ION | | Authors: | Woo, L.W.L, Jackson, T, Putey, A, Cozier, G, Leonard, P, Acharya, K.R, Chander, S.K, Purohit, A, Reed, M.J, Potter, B.V.L. | | Deposit date: | 2009-03-19 | | Release date: | 2010-02-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Highly Potent First Examples of Dual Aromatase-Steroid Sulfatase Inhibitors Based on a Biphenyl Template.
J.Med.Chem., 53, 2010
|
|
2WA8
 
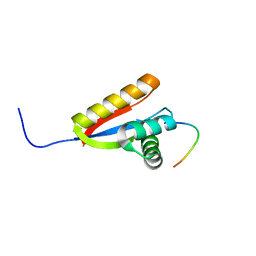 | | Structural basis of N-end rule substrate recognition in Escherichia coli by the ClpAP adaptor protein ClpS - The Phe peptide structure | | Descriptor: | ATP-DEPENDENT CLP PROTEASE ADAPTER PROTEIN CLPS, N-END RULE PEPTIDE | | Authors: | Schuenemann, V.J, Kralik, S.M, Albrecht, R, Spall, S.K, Truscott, K.N, Dougan, D.A, Zeth, K. | | Deposit date: | 2009-02-03 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis of N-End Rule Substrate Recognition in Escherichia Coli by the Clpap Adaptor Protein Clps.
Embo Rep., 10, 2009
|
|
2W8M
 
 | |
2XEH
 
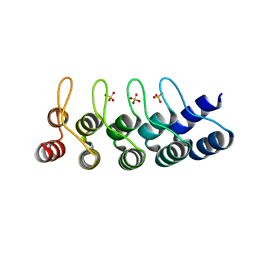 | | Structural Determinants for Improved Thermal Stability of Designed Ankyrin Repeat Proteins With a Redesigned C-capping Module. | | Descriptor: | NI3C MUT6, SULFATE ION | | Authors: | Kramer, M, Wetzel, S.K, Pluckthun, A, Mittl, P, Grutter, M. | | Deposit date: | 2010-05-14 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Determinants for Improved Thermal Stability of Designed Ankyrin Repeat Proteins with a Redesigned C-Capping Module.
J.Mol.Biol., 404, 2010
|
|
2XEE
 
 | | Structural Determinants for Improved Thermal Stability of Designed Ankyrin Repeat Proteins With a Redesigned C-capping Module. | | Descriptor: | NI3C DARPIN MUTANT5 | | Authors: | Kramer, M, Wetzel, S.K, Pluckthun, A, Mittl, P, Grutter, M. | | Deposit date: | 2010-05-14 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Determinants for Improved Thermal Stability of Designed Ankyrin Repeat Proteins with a Redesigned C-Capping Module.
J.Mol.Biol., 404, 2010
|
|
2WA9
 
 | | Structural basis of N-end rule substrate recognition in Escherichia coli by the ClpAP adaptor protein ClpS - Trp peptide structure | | Descriptor: | ATP-DEPENDENT CLP PROTEASE ADAPTER PROTEIN CLPS, TRP PEPTIDE | | Authors: | Schuenemann, V.J, Kralik, S.M, Albrecht, R, Spall, S.K, Truscott, K.N, Dougan, D.A, Zeth, K. | | Deposit date: | 2009-02-03 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis of N-End Rule Substrate Recognition in Escherichia Coli by the Clpap Adaptor Protein Clps.
Embo Rep., 10, 2009
|
|
5FWQ
 
 | | Apo structure of human Leukotriene A4 hydrolase | | Descriptor: | ACETATE ION, HUMAN LEUKOTRIENE A4 HYDROLASE, IMIDAZOLE, ... | | Authors: | Wittmann, S.K, Kalinowsky, L, Kramer, J, Bloecher, R, Steinhilber, D, Pogoryelov, D, Proschak, E, Heering, J. | | Deposit date: | 2016-02-19 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Thermodynamic properties of leukotriene A4hydrolase inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
2WLS
 
 | | Crystal structure of Mus musculus Acetylcholinesterase in complex with AMTS13 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pang, Y.P, Ekstrom, F, Polsinelli, G.A, Gao, Y, Rana, S, Hua, D.H, Andersson, B, Andersson, P.O, Peng, L, Singh, S.K, Mishra, R.K, Zhu, K.Y, Fallon, A.M, Ragsdale, D.W, Brimijoin, S. | | Deposit date: | 2009-06-25 | | Release date: | 2009-09-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Selective and Irreversible Inhibitors of Mosquito Acetylcholinesterases for Controlling Malaria and Other Mosquito-Borne Diseases.
Plos One, 4, 2009
|
|
2VNZ
 
 | | Crystal structure of dithinonite reduced soybean ascorbate peroxidase mutant W41A. | | Descriptor: | ASCORBATE PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION | | Authors: | Metcalfe, C.L, Badyal, S.K, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Iron Oxidation State Modulates Active Site Structure in a Heme Peroxidase.
Biochemistry, 47, 2008
|
|
2VNX
 
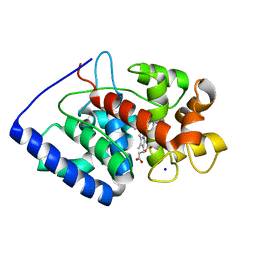 | | Crystal structure of soybean ascorbate peroxidase mutant W41A after exposure to a high dose of x-rays | | Descriptor: | ASCORBATE PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION | | Authors: | Metcalfe, C.L, Badyal, S.K, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Iron Oxidation State Modulates Active Site Structure in a Heme Peroxidase.
Biochemistry, 47, 2008
|
|
2WD4
 
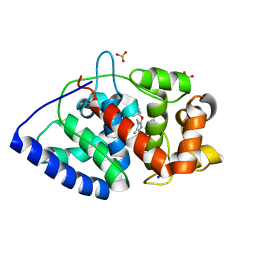 | | Ascorbate Peroxidase as a heme oxygenase: w41A variant product with t-butyl peroxide | | Descriptor: | 3-[2-[[3-(2-CARBOXYETHYL)-5-[[3-ETHENYL-4-METHYL-5-[(2-METHYLPROPAN-2-YL)OXY]-1H-PYRROL-2-YL]METHYL]-4-METHYL-1H-PYRROL -2-YL]METHYL]-5-[(Z)-(4-ETHENYL-3-METHYL-5-OXO-PYRROL-2-YLIDENE)METHYL]-4-METHYL-1H-PYRROL-3-YL]PROPANOIC ACID, ASCORBATE PEROXIDASE, FE (III) ION, ... | | Authors: | Badyal, S.K, Metcalfe, C.L, Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2009-03-19 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evidence for Heme Oxygenase Activity in a Heme Peroxidase.
Biochemistry, 48, 2009
|
|
2XTH
 
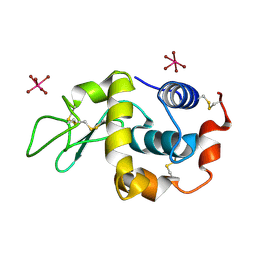 | | K2PtBr6 binding to lysozyme | | Descriptor: | HEXABROMOPLATINATE(IV), LYSOZYME C | | Authors: | Helliwell, J.R, Bell, A.M.T, Bryant, P, Fisher, S, Habash, J, Helliwell, M, Margiolaki, I, Kaenket, S, Watier, Y, Wright, J, Yalamanchili, S.K. | | Deposit date: | 2010-10-07 | | Release date: | 2010-12-08 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Time-Dependent Analysis of K2Ptbr6 Binding to Lysozyme Studied by Protein Powder and Single Crystal X-Ray Analysis
Z.Kristallogr., 225, 2010
|
|
2XEN
 
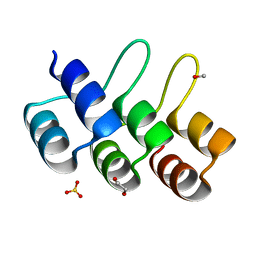 | | Structural Determinants for Improved Thermal Stability of Designed Ankyrin Repeat Proteins With a Redesigned C-capping Module. | | Descriptor: | 1,2-ETHANEDIOL, METHANOL, NI1C MUT4, ... | | Authors: | Kramer, M, Wetzel, S.K, Pluckthun, A, Mittl, P, Grutter, M. | | Deposit date: | 2010-05-17 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Determinants for Improved Thermal Stability of Designed Ankyrin Repeat Proteins with a Redesigned C-Capping Module.
J.Mol.Biol., 404, 2010
|
|
2YBD
 
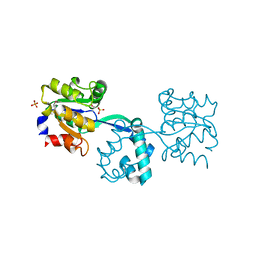 | | Crystal structure of probable had family hydrolase from pseudomonas fluorescens pf-5 with bound phosphate | | Descriptor: | HYDROLASE, HALOACID DEHALOGENASE-LIKE FAMILY, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Dunaway-Mariano, D, Allen, K.N, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-03-03 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Probable Had Family Hydrolase from Pseudomonas Fluorescens Pf-5 with Bound Phosphate
To be Published
|
|
1J98
 
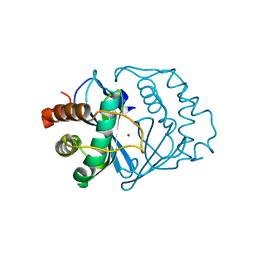 | | The 1.2 Angstrom Structure of Bacillus subtilis LuxS | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, ZINC ION | | Authors: | Ruzheinikov, S.N, Das, S.K, Sedelnikova, S.E, Hartley, A, Foster, S.J, Horsburgh, M.J, Cox, A.G, McCleod, C.W, Mekhalfia, A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-05-24 | | Release date: | 2001-06-06 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2 A Structure of a Novel Quorum-Sensing Protein, Bacillus subtilis LuxS
J.Mol.Biol., 313, 2001
|
|
1J2X
 
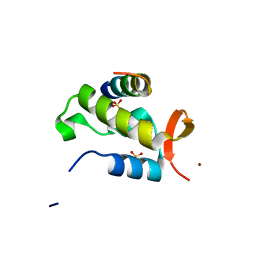 | | Crystal structure of RAP74 C-terminal domain complexed with FCP1 C-terminal peptide | | Descriptor: | RNA polymerase II CTD phosphatase, SULFATE ION, Transcription initiation factor IIF, ... | | Authors: | Kamada, K, Roeder, R.G, Burley, S.K. | | Deposit date: | 2003-01-15 | | Release date: | 2003-01-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular mechanism of recruitment of TFIIF- associating RNA polymerase C-terminal domain phosphatase (FCP1) by transcription factor IIF
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
8OYP
 
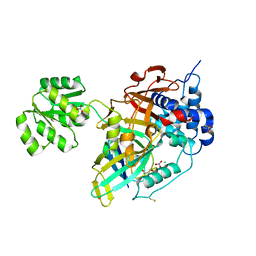 | | Crystal structure of Ubiquitin specific protease 11 (USP11) in complex with a substrate mimetic | | Descriptor: | CADMIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Maurer, S.K, Caulton, S.G, Ward, S.J, Emsley, J, Dreveny, I. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Ubiquitin-specific protease 11 structure in complex with an engineered substrate mimetic reveals a molecular feature for deubiquitination selectivity.
J.Biol.Chem., 299, 2023
|
|
8PXS
 
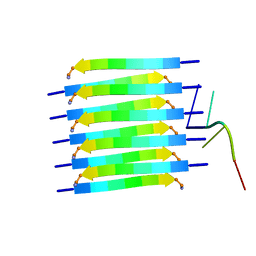 | | Short RNA binding to peptide amyloids | | Descriptor: | RNA (5'-R(P*GP*UP*CP*A)-3'), VAL-ALA-GLN-ALA-GLN-ILE-ASN-ILE | | Authors: | Rout, S.K, Cadalbert, R, Schroder, N, Wiegand, T, Zehnder, J, Gampp, O, Guntert, P, Kringler, D, Kreutz, C, Knorlein, A, Hall, J, Greenwald, J, Riek, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-16 | | Method: | SOLID-STATE NMR | | Cite: | An Analysis of Nucleotide-Amyloid Interactions Reveals Selective Binding to Codon-Sized RNA.
J.Am.Chem.Soc., 145, 2023
|
|
8Q6K
 
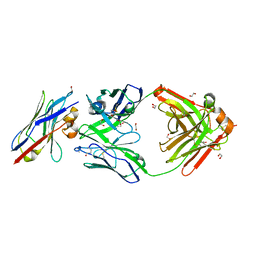 | | Human IgD Fab in complex with an orthosteric inhibitor of Phl p 7 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Human IgD Fab heavy chain, ... | | Authors: | Davies, A.M, Vester, S.K, McDonnell, J.M. | | Deposit date: | 2023-08-11 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Expanding the Anti-Phl p 7 Antibody Toolkit: An Anti-Idiotype Nanobody Inhibitor.
Antibodies, 12, 2023
|
|
8Q52
 
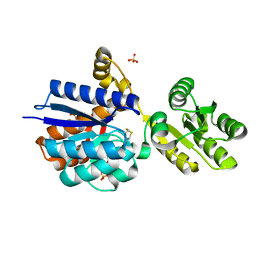 | | A PBP-like protein built from fragments of different folds | | Descriptor: | Leucine-specific-binding protein,Chemotaxis protein CheY, SULFATE ION | | Authors: | Shanmugaratnam, S, Toledo-Patino, S, Goetz, S.K, Farias-Rico, J.A, Hocker, B. | | Deposit date: | 2023-08-08 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular handcraft of a well-folded protein chimera.
Febs Lett., 598, 2024
|
|
8JOJ
 
 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 9,11-epoxy-17-hydroxypregn-4-ene-3,20-dione actate | | Descriptor: | 3-ketosteroid dehydrogenase, 9,11-epoxy-17-hydroxypregn-4-ene-3,20-dione actate, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hu, Y.L, Li, X, Cheng, X.Y, Song, S.K, Su, Z.D. | | Deposit date: | 2023-06-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.819 Å) | | Cite: | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 9,11-epoxy-17-hydroxypregn-4-ene-3,20-dione actate
To Be Published
|
|
8KCZ
 
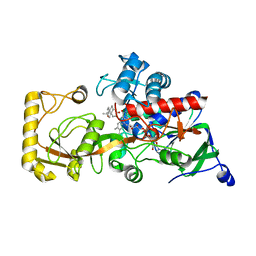 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 1,4-androstadiene-3,17- dione | | Descriptor: | 3-ketosteroid dehydrogenase, ANDROSTA-1,4-DIENE-3,17-DIONE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hu, Y.L, Li, X, Cheng, X.Y, Song, S.K, Su, Z.D. | | Deposit date: | 2023-08-08 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 1,4-androstadiene-3,17- dione
To Be Published
|
|
8KC6
 
 | | X-ray structure of 3-alpha-(Or 20-beta)-hydroxysteroid dehydrogenase | | Descriptor: | 3-alpha-(Or 20-beta)-hydroxysteroid dehydrogenase | | Authors: | Song, S.K, Lv, D.Q, Cheng, X.Y, Ma, Q.Q, Hu, Y.L, Su, Z.D. | | Deposit date: | 2023-08-06 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | X-ray structure of 3-alpha-(Or 20-beta)-hydroxysteroid dehydrogenase
To Be Published
|
|
8JU4
 
 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 1,4-androstadiene-3,17- dione | | Descriptor: | 3-ketosteroid dehydrogenase, ANDROSTA-1,4-DIENE-3,17-DIONE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hu, Y.L, Li, X, Cheng, X.Y, Song, S.K, Su, Z.D. | | Deposit date: | 2023-06-24 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 1,4-androstadiene-3,17- dione
To Be Published
|
|
8KCN
 
 | |
