5KUJ
 
 | | Human mitochondrial calcium uniporter (residues 72-189) crystal structure with magnesium. | | Descriptor: | Calcium uniporter protein, mitochondrial, MAGNESIUM ION | | Authors: | Mok, M.C.Y, Lee, S.K, Junop, M.S, Stathopulos, P.B. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights into Mitochondrial Calcium Uniporter Regulation by Divalent Cations.
Cell Chem Biol, 23, 2016
|
|
5VND
 
 | | Crystal structure of FGFR1-Y563C (FGFR4 surrogate) covalently bound to H3B-6527 | | Descriptor: | 1,2-ETHANEDIOL, Fibroblast growth factor receptor 1, N-{2-[(6-{[(2,6-dichloro-3,5-dimethoxyphenyl)carbamoyl](methyl)amino}pyrimidin-4-yl)amino]-5-(4-ethylpiperazin-1-yl)phenyl}propanamide, ... | | Authors: | Tsai, J.H.C, Reynolds, D, Fekkes, P, Smith, P, Larsen, N.A. | | Deposit date: | 2017-04-30 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | H3B-6527 Is a Potent and Selective Inhibitor of FGFR4 in FGF19-Driven Hepatocellular Carcinoma.
Cancer Res., 77, 2017
|
|
7UBO
 
 | | Crystal Structure of the first bromodomain of human BRDT in complex with the inhibitor CCD-956 | | Descriptor: | Bromodomain testis-specific protein, DIMETHYL SULFOXIDE, N-[(2R)-1-(methylamino)-3-{1-[(4-methyl-2-oxo-1,2-dihydroquinolin-6-yl)acetyl]piperidin-4-yl}-1-oxopropan-2-yl]-5-phenylpyridine-2-carboxamide | | Authors: | Ta, H.M, Modukuri, R.K, Yu, Z, Tan, Z, Matzuk, M.M, Kim, C. | | Deposit date: | 2022-03-15 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery of potent BET bromodomain 1 stereoselective inhibitors using DNA-encoded chemical library selections.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1V3W
 
 | | Structure of Ferripyochelin binding protein from Pyrococcus horikoshii OT3 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Jeyakanthan, J, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-07 | | Release date: | 2003-11-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Observation of a calcium-binding site in the gamma-class carbonic anhydrase from Pyrococcus horikoshii.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1V67
 
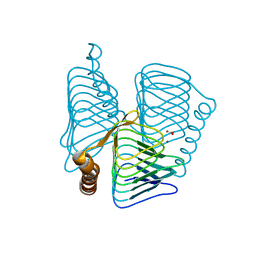 | | Structure of ferripyochelin binding protein from pyrococcus horikoshii OT3 | | Descriptor: | BICARBONATE ION, CALCIUM ION, ZINC ION, ... | | Authors: | Jeyakanthan, J, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-27 | | Release date: | 2003-12-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Observation of a calcium-binding site in the gamma-class carbonic anhydrase from Pyrococcus horikoshii.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
6GRE
 
 | |
6GRF
 
 | |
2FKO
 
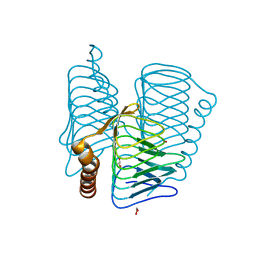 | | Structure of PH1591 from Pyrococcus horikoshii OT3 | | Descriptor: | 1,2-ETHANEDIOL, 173aa long hypothetical ferripyochelin binding protein, ZINC ION | | Authors: | Jeyakanthan, J, Tahirov, T.H, Yokoyama, S, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-01-05 | | Release date: | 2007-01-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Observation of a calcium-binding site in the gamma-class carbonic anhydrase from Pyrococcus horikoshii.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
5XGD
 
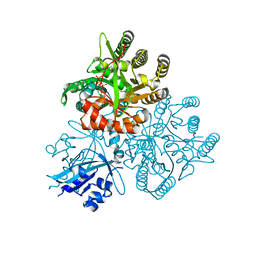 | | Crystal structure of the PAS-GGDEF-EAL domain of PA0861 from Pseudomonas aeruginosa in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Uncharacterized protein PA0861 | | Authors: | Liu, C, Liew, C.W, Sreekanth, R, Lescar, J. | | Deposit date: | 2017-04-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into Biofilm Dispersal Regulation from the Crystal Structure of the PAS-GGDEF-EAL Region of RbdA from Pseudomonas aeruginosa.
J. Bacteriol., 200, 2018
|
|
8T4Z
 
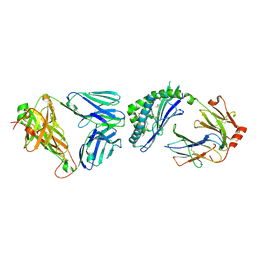 | | T-cell receptor and lipid complex structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, Beta-2-microglobulin, ... | | Authors: | Praveena, T, Rossjohn, J. | | Deposit date: | 2023-06-12 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Lipidomic scanning of self-lipids identifies headless antigens for natural killer T cells.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6AC9
 
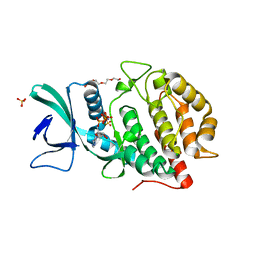 | | Crystal structure of human Vaccinia-related kinase 1 (VRK1) in complex with AMP-PNP | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Ngow, Y.S, Sreekanth, R, Yoon, H.S. | | Deposit date: | 2018-07-25 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of human vaccinia-related kinase 1 in complex with AMP-PNP, a non-hydrolyzable ATP analog.
Protein Sci., 28, 2019
|
|
5XGE
 
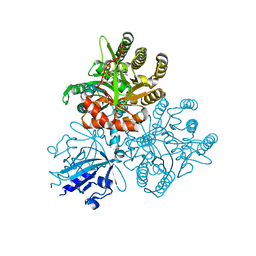 | | Crystal structure of the PAS-GGDEF-EAL domain of PA0861 from Pseudomonas aeruginosa in complex with cyclic di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Uncharacterized protein PA0861 | | Authors: | Liu, C, Liew, C.W, Sreekanth, R, Lescar, J. | | Deposit date: | 2017-04-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Insights into Biofilm Dispersal Regulation from the Crystal Structure of the PAS-GGDEF-EAL Region of RbdA from Pseudomonas aeruginosa.
J. Bacteriol., 200, 2018
|
|
3NI6
 
 | |
5XGB
 
 | |
5C39
 
 | |
4ITZ
 
 | |
4J4O
 
 | | Crystal structure of FK506 binding domain of plasmodium VIVAX FKBP35 in complex with D44 | | Descriptor: | 70 kDa peptidylprolyl isomerase, putative, GLYCEROL, ... | | Authors: | Sreekanth, R, Harikishore, A, Yoon, H.S. | | Deposit date: | 2013-02-07 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Small molecule Plasmodium FKBP35 inhibitor as a potential antimalaria agent.
Sci Rep, 3, 2013
|
|
4J4N
 
 | |
7L99
 
 | | Crystal structure of BRDT bromodomain 2 in complex with CDD-1302 | | Descriptor: | Bromodomain testis-specific protein, N-[3-(acetylamino)-4-methylphenyl]-3-(4-amino-2-methylphenyl)-1-methyl-1H-indazole-5-carboxamide, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500) | | Authors: | Sharma, R, Yu, Z, Ku, A.F, Anglin, J.L, Ucisik, M.N, Faver, J.C, Sankaran, B, Kim, C, Matzuk, M.M. | | Deposit date: | 2021-01-03 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and characterization of bromodomain 2-specific inhibitors of BRDT.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7L9A
 
 | | Crystal structure of BRDT bromodomain 2 in complex with CDD-1102 | | Descriptor: | BETA-MERCAPTOETHANOL, Bromodomain testis-specific protein, N~1~-(5-{[3-(4-amino-2-methylphenyl)-1-methyl-1H-indazole-5-carbonyl]amino}-2-methylphenyl)-N~4~-methylbenzene-1,4-dicarboxamide | | Authors: | Sharma, R, Kaur, G, Yu, Z, Ku, A.F, Anglin, J.L, Ucisik, M.N, Faver, J.C, Sankaran, B, Kim, C, Matzuk, M.M. | | Deposit date: | 2021-01-03 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery and characterization of bromodomain 2-specific inhibitors of BRDT.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4F2H
 
 | |
6C0A
 
 | | Actinin-1 EF-Hand bound to the Cav1.2 IQ Motif | | Descriptor: | Alpha-actinin-1, Voltage-dependent L-type calcium channel subunit alpha-1C | | Authors: | Turner, M.L, Ames, J.B. | | Deposit date: | 2017-12-28 | | Release date: | 2019-01-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Localization and Activation of Neuronal L-type Ca2+ Channels by a-Actinin1 and Calmodulin
To be Published
|
|
3PA7
 
 | | Crystal structure of FKBP from plasmodium vivax in complex with tetrapeptide ALPF | | Descriptor: | 4-mer Peptide ALPF, 70 kDa peptidylprolyl isomerase, putative | | Authors: | Balakrishna, A.M, Alag, R, Yoon, H.S. | | Deposit date: | 2010-10-18 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural insights into substrate binding by PvFKBP35, a peptidylprolyl cis-trans isomerase from the human malarial parasite Plasmodium vivax
EUKARYOTIC CELL, 12, 2013
|
|
2MPH
 
 | |
2WQJ
 
 | |
