7SZQ
 
 | | Human P300 complexed with an azaindazole inhibitor | | Descriptor: | 1-[1-(4-chlorophenyl)cyclopentane-1-carbonyl]-N-1H-pyrazolo[4,3-b]pyridin-5-yl-D-prolinamide, Histone acetyltransferase p300 | | Authors: | Shewchuk, L.M, Reid, R.A. | | Deposit date: | 2021-11-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Proline-Based p300/CBP Inhibitors Using DNA-Encoded Library Technology in Combination with High-Throughput Screening.
J.Med.Chem., 65, 2022
|
|
7SSK
 
 | | Human P300 complexed with a glycine-based inhibitor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Histone acetyltransferase p300, ... | | Authors: | Shewchuk, L.M, Reid, R.A. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Discovery of Proline-Based p300/CBP Inhibitors Using DNA-Encoded Library Technology in Combination with High-Throughput Screening.
J.Med.Chem., 65, 2022
|
|
7SS8
 
 | | Human P300 complexed with a proline-based inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 1-[1-(4-chlorophenyl)cyclopentane-1-carbonyl]-N-{[3-(methylcarbamoyl)phenyl]methyl}-D-prolinamide, Histone acetyltransferase p300, ... | | Authors: | Shewchuk, L.M, Reid, R.A. | | Deposit date: | 2021-11-10 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of Proline-Based p300/CBP Inhibitors Using DNA-Encoded Library Technology in Combination with High-Throughput Screening.
J.Med.Chem., 65, 2022
|
|
7B2B
 
 | | Solution structure of a non-covalent extended docking domain complex of the Pax NRPS: PaxA T1-CDD/PaxB NDD | | Descriptor: | Amino acid adenylation domain-containing protein, Peptide synthetase PaxA | | Authors: | Watzel, J, Sarawi, S, Duchardt-Ferner, E, Bode, H.B, Woehnert, J. | | Deposit date: | 2020-11-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Cooperation between a T Domain and a Minimal C-Terminal Docking Domain to Enable Specific Assembly in a Multiprotein NRPS.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7B2F
 
 | | Solution structure of the Pax NRPS docking domain PaxB NDD | | Descriptor: | Peptide synthetase XpsB (Modular protein) | | Authors: | Watzel, J, Sarawi, S, Duchardt-Ferner, E, Bode, H.B, Woehnert, J. | | Deposit date: | 2020-11-26 | | Release date: | 2021-06-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Cooperation between a T Domain and a Minimal C-Terminal Docking Domain to Enable Specific Assembly in a Multiprotein NRPS.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6FTR
 
 | | Serial Femtosecond Crystallography at Megahertz pulse rates | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wiedorn, M.O, Oberthuer, D, Barty, A, Chapman, H.N. | | Deposit date: | 2018-02-23 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76000106 Å) | | Cite: | Megahertz serial crystallography.
Nat Commun, 9, 2018
|
|
6GTH
 
 | | Serial Femtosecond Crystallography at Megahertz pulse rates | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Wiedorn, M, Oberthuer, D, Werner, N, Schubert, R, White, T.A, Mancuso, A, Perbandt, M, Betzel, C, Barty, A, Chapman, H. | | Deposit date: | 2018-06-18 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Megahertz serial crystallography.
Nat Commun, 9, 2018
|
|
7ZOC
 
 | |
487D
 
 | |
8Q7Y
 
 | |
3ELM
 
 | | Crystal Structure of MMP-13 Complexed with Inhibitor 24f | | Descriptor: | (2R)-({[5-(4-ethoxyphenyl)thiophen-2-yl]sulfonyl}amino){1-[(1-methylethoxy)carbonyl]piperidin-4-yl}ethanoic acid, CALCIUM ION, Collagenase 3, ... | | Authors: | Kulathila, R, Monovich, L, Koehn, J. | | Deposit date: | 2008-09-22 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of potent, selective, and orally active carboxylic acid based inhibitors of matrix metalloproteinase-13
J.Med.Chem., 52, 2009
|
|
5MBW
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH Pep#3 | | Descriptor: | BACE1 INHIBITOR PEPTIDE Pep#3, Beta-secretase 1, CHLORIDE ION | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-09 | | Release date: | 2017-09-27 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Potent and Selective BACE-1 Peptide Inhibitors Lower Brain A beta Levels Mediated by Brain Shuttle Transport.
EBioMedicine, 24, 2017
|
|
5MCO
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH ACTIVE SITE INHIBITOR GRL-8234 AND EXOSITE PEPTIDE | | Descriptor: | BACE-1 EXOSITE PEPTIDE, Beta-secretase 1, N-{(1S,2R)-1-benzyl-2-hydroxy-3-[(3-methoxybenzyl)amino]propyl}-5-[methyl(methylsulfonyl)amino]-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-09-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Potent and Selective BACE-1 Peptide Inhibitors Lower Brain A beta Levels Mediated by Brain Shuttle Transport.
EBioMedicine, 24, 2017
|
|
5MCQ
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH ACTIVE SITE AND EXOSITE BINDING PEPTIDE INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, BACE-1 ACTIVE AND EXOSITE BINDING INHIBITOR, Beta-secretase 1 | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-09-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Potent and Selective BACE-1 Peptide Inhibitors Lower Brain A beta Levels Mediated by Brain Shuttle Transport.
EBioMedicine, 24, 2017
|
|
6TET
 
 | | The structure of CYP121 in complex with inhibitor L21 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(~{E})-3-[4-(4-fluorophenyl)phenyl]prop-2-enyl]imidazole, Mycocyclosin synthase, ... | | Authors: | Adam, S, Koehnke, J. | | Deposit date: | 2019-11-12 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49986887 Å) | | Cite: | Structure-Activity Relationship and Mode-Of-Action Studies Highlight 1-(4-Biphenylylmethyl)-1H-imidazole-Derived Small Molecules as Potent CYP121 Inhibitors.
Chemmedchem, 16, 2021
|
|
6TEV
 
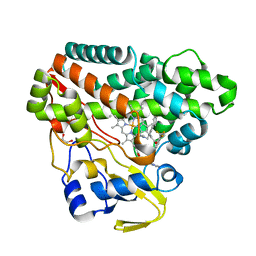 | | The structure of CYP121 in complex with inhibitor L44 | | Descriptor: | 1,2-ETHANEDIOL, 1-[[4-[4-(trifluoromethyl)phenyl]phenyl]methyl]imidazole, Mycocyclosin synthase, ... | | Authors: | Adam, S, Koehnke, J. | | Deposit date: | 2019-11-12 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.70001268 Å) | | Cite: | Structure-Activity Relationship and Mode-Of-Action Studies Highlight 1-(4-Biphenylylmethyl)-1H-imidazole-Derived Small Molecules as Potent CYP121 Inhibitors.
Chemmedchem, 16, 2021
|
|
6TE7
 
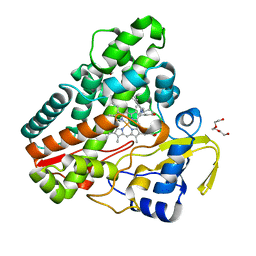 | | The structure of CYP121 in complex with inhibitor S2 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[4-[(1~{R})-1-imidazol-1-ylprop-2-enyl]phenyl]phenol, Mycocyclosin synthase, ... | | Authors: | Adam, S, Koehnke, J. | | Deposit date: | 2019-11-11 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.50001824 Å) | | Cite: | Structure-Activity Relationship and Mode-Of-Action Studies Highlight 1-(4-Biphenylylmethyl)-1H-imidazole-Derived Small Molecules as Potent CYP121 Inhibitors.
Chemmedchem, 16, 2021
|
|
3RKF
 
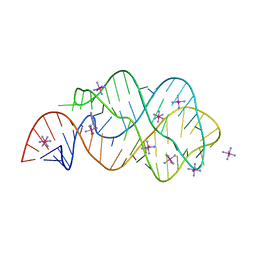 | | Crystal structure of guanine riboswitch C61U/G37A double mutant bound to thio-guanine | | Descriptor: | 2-amino-1,9-dihydro-6H-purine-6-thione, COBALT HEXAMMINE(III), Guanine riboswitch | | Authors: | Buck, J, Wacker, A, Warkentin, E, Woehnert, J, Wirmer-Bartoschek, J, Schwalbe, H. | | Deposit date: | 2011-04-18 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Influence of ground-state structure and Mg2+ binding on folding kinetics of the guanine-sensing riboswitch aptamer domain.
Nucleic Acids Res., 39, 2011
|
|
1A4B
 
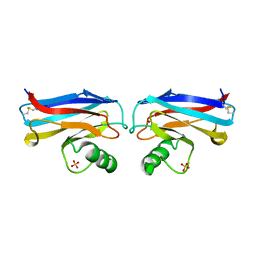 | |
1A4C
 
 | | AZURIN MUTANT WITH MET 121 REPLACED BY HIS, PH 3.5 CRYSTAL FORM, DATA COLLECTED AT-180 DEGREES CELSIUS | | Descriptor: | AZURIN, COPPER (II) ION, NITRATE ION, ... | | Authors: | Messerschmidt, A, Prade, L. | | Deposit date: | 1998-01-28 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Rack-induced metal binding vs. flexibility: Met121His azurin crystal structures at different pH.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1A4A
 
 | |
1B75
 
 | |
6S10
 
 | |
8GBX
 
 | |
8GBY
 
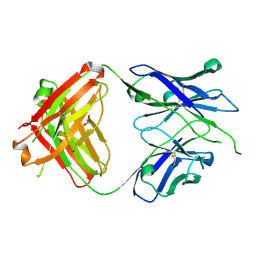 | | Crystal structure of PC39-50E, an anti-HIV broadly neutralizing antibody | | Descriptor: | GLYCEROL, PC39-50E Fab heavy chain, PC39-50E Fab light chain | | Authors: | Murrell, S, Omorodion, O, Wilson, I.A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Antigen pressure from two founder viruses induces multiple insertions at a single antibody position to generate broadly neutralizing HIV antibodies.
Plos Pathog., 19, 2023
|
|
