5ZKL
 
 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT12 | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), SP_0782 | | Authors: | Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2018-03-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
5ZKM
 
 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA TCTTCC | | Descriptor: | DNA (5'-D(P*TP*CP*TP*TP*CP*C)-3'), SP_0782 | | Authors: | Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2018-03-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
7DPG
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle | | Descriptor: | VP2, VP3, Virion protein 1 | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q, Xia, N. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ7
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 5F5 | | Descriptor: | 5F5 VH, 5F5 VL, Capsid protein VP4, ... | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ4
 
 | | Cryo-EM structure of CAR triggered Coxsackievirus B1 A-particle | | Descriptor: | VP2, VP3, Virion protein 1 | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPZ
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR | | Descriptor: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ1
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR at physiological temperature | | Descriptor: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
1QWK
 
 | | Structural genomics of Caenorhabditis Elegans: Hypothetical 35.2 kDa protein (aldose reductase family member) | | Descriptor: | aldo-keto reductase family 1 member C1 | | Authors: | Chen, L, Zhou, X.E, Meehan, E.J, Symersky, J, Lu, S, Li, S, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-09-02 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural genomics of Caenorhabditis Elegans: Hypothetical 35.2 kDa
protein (aldose reductase family member)
To be published
|
|
5WSO
 
 | |
2AHX
 
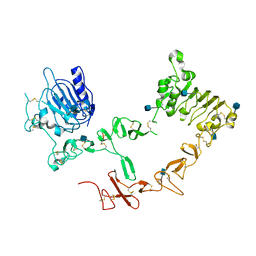 | | Crystal structure of ErbB4/HER4 extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor tyrosine-protein kinase erbB-4, SULFATE ION, ... | | Authors: | Bouyain, S, Longo, P.A, Li, S, Ferguson, K.M, Leahy, D.J. | | Deposit date: | 2005-07-28 | | Release date: | 2005-09-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | The extracellular region of ErbB4 adopts a tethered conformation in the absence of ligand
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
4MY5
 
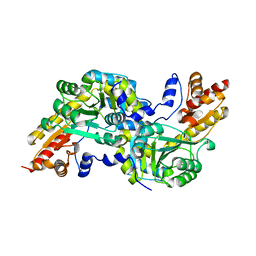 | | Crystal structure of the aromatic amino acid aminotransferase from Streptococcus mutants | | Descriptor: | Putative amino acid aminotransferase | | Authors: | Cong, X, Li, X, Ge, J, Feng, Y, Feng, X, Li, S. | | Deposit date: | 2013-09-27 | | Release date: | 2014-10-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Crystal structure of the aromatic-amino-acid aminotransferase from Streptococcus mutans.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6E3S
 
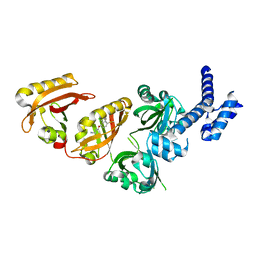 | | Crystal Structure of the Heterodimeric HIF-2 Complex with Antagonist PT2385 | | Descriptor: | 3-{[(1S)-2,2-difluoro-1-hydroxy-7-(methylsulfonyl)-2,3-dihydro-1H-inden-4-yl]oxy}-5-fluorobenzonitrile, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Wu, D, Su, X, Lu, J, Li, S, Hood, B, Vasile, S, Potluri, N, Diao, X, Kim, Y, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2018-07-15 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bidirectional modulation of HIF-2 activity through chemical ligands.
Nat. Chem. Biol., 15, 2019
|
|
6E3T
 
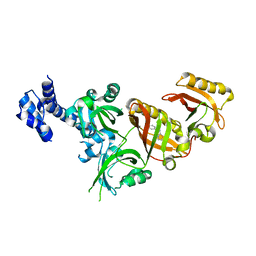 | | Crystal Structure of the Heterodimeric HIF-2 Complex with Antagonist T1001 | | Descriptor: | (6S)-6-(4-bromophenyl)-2,3,5,6-tetrahydroimidazo[2,1-b][1,3]thiazole, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Wu, D, Su, X, Lu, J, Li, S, Hood, B, Vasile, S, Potluri, N, Diao, X, Kim, Y, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2018-07-15 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bidirectional modulation of HIF-2 activity through chemical ligands.
Nat. Chem. Biol., 15, 2019
|
|
6E3U
 
 | | Crystal Structure of the Heterodimeric HIF-2 Complex with Agonist M1001 | | Descriptor: | 3-{[2-(pyrrolidin-1-yl)phenyl]amino}-1H-1lambda~6~,2-benzothiazole-1,1-dione, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Wu, D, Su, X, Lu, J, Li, S, Hood, B, Vasile, S, Potluri, N, Diao, X, Kim, Y, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2018-07-15 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Bidirectional modulation of HIF-2 activity through chemical ligands.
Nat. Chem. Biol., 15, 2019
|
|
7XN4
 
 | | Cryo-EM structure of CopC-CaM-caspase-3 with NAD+ | | Descriptor: | Arginine ADP-riboxanase CopC, Calmodulin-1, Caspase-3, ... | | Authors: | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
7XN5
 
 | | Cryo-EM structure of CopC-CaM-caspase-3 with ADPR | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Arginine ADP-riboxanase CopC, Calmodulin-1, ... | | Authors: | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
7XN6
 
 | | Cryo-EM structure of CopC-CaM-caspase-3 with ADPR-deacylization | | Descriptor: | Arginine ADP-riboxanase CopC, Calmodulin-1, Caspase-3, ... | | Authors: | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
5WPR
 
 | | Crystal structure HpiC1 in C2 space group | | Descriptor: | 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
5WPP
 
 | | Crystal structure HpiC1 W73M/K132M | | Descriptor: | 12-epi-hapalindole C/U synthase, CALCIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
5WPU
 
 | | Crystal structure HpiC1 Y101S | | Descriptor: | 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
5WPS
 
 | | Crystal structure HpiC1 Y101F | | Descriptor: | 1,2-ETHANEDIOL, 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.389 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
3KS4
 
 | | Crystal structure of Reston ebolavirus VP35 RNA binding domain | | Descriptor: | Polymerase cofactor VP35 | | Authors: | Kimberlin, C.R, Bornholdt, Z.A, Li, S, Woods, V.L, Macrae, I.J, Saphire, E.O. | | Deposit date: | 2009-11-20 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression.
Proc.Natl.Acad.Sci.USA, 107, 2009
|
|
5OPG
 
 | | Structure of the Hantaan virus Gn glycoprotein ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Rissanen, I, Stass, R, Zeltina, A, Li, S, Hepojoki, J, Harlos, K, Gilbert, R.J.C, Huiskonen, J.T, Bowden, T.A. | | Deposit date: | 2017-08-09 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Transitions of the Conserved and Metastable Hantaviral Glycoprotein Envelope.
J. Virol., 91, 2017
|
|
6LB0
 
 | |
7X7T
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zheng, Q, Li, S, Zhang, T, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
