8FU4
 
 | | HCMV US11 peptide binding to HLA-A*02:01 | | Descriptor: | Beta-2-microglobulin, CADMIUM ION, GLYCEROL, ... | | Authors: | Watson, G.M, Berry, R, Littler, D.R, Rossjohn, J. | | Deposit date: | 2023-01-16 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Diverse cytomegalovirus US11 antagonism and MHC-A evasion strategies reveal a tit-for-tat coevolutionary arms race in hominids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8FRT
 
 | | X-ray crystal structure of the N-terminal region from HCMV US11 binding to HLA-A*02:01 | | Descriptor: | GLYCEROL, Unique short US11 glycoprotein, Beta-2-microglobulin, ... | | Authors: | Watson, G.M, Berry, R, Rossjohn, J. | | Deposit date: | 2023-01-08 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Diverse cytomegalovirus US11 antagonism and MHC-A evasion strategies reveal a tit-for-tat coevolutionary arms race in hominids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5KU9
 
 | | Crystal structure of MCL1 with compound 1 | | Descriptor: | (3~{S})-3-azanyl-4-(4-bromophenyl)-~{N}-[(3~{S})-1-[2-[[(2~{R})-1-(3,4-dichlorophenyl)-4-(methylamino)-4-oxidanylidene-butan-2-yl]amino]-2-oxidanylidene-ethyl]-2-oxidanylidene-4,5-dihydro-3~{H}-1-benzazepin-3-yl]butanamide, Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION | | Authors: | Ferguson, A.D. | | Deposit date: | 2016-07-13 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
5MES
 
 | | MCL1 FAB COMPLEX IN COMPLEX WITH COMPOUND 29 | | Descriptor: | (5~{R},13~{S},17~{S})-5-[[4-chloranyl-3-(2-phenylethyl)phenyl]methyl]-13-[(4-chlorophenyl)methyl]-8-methyl-1,4,8,12,16-pentazatricyclo[15.8.1.0^{20,25}]hexacosa-20(25),21,23-triene-3,7,15,26-tetrone, Heavy Chain, Induced myeloid leukemia cell differentiation protein Mcl-1 homolog,Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Hargreaves, D. | | Deposit date: | 2016-11-16 | | Release date: | 2017-01-18 | | Last modified: | 2017-03-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
5MEV
 
 | | MCL1 FAB COMPLEX IN COMPLEX WITH COMPOUND 21 | | Descriptor: | (5~{R},13~{S},17~{S})-5-[(3,4-dichlorophenyl)methyl]-8-methyl-13-[(4-methylsulfonylphenyl)methyl]-1,4,8,12,16-pentazatricyclo[15.8.1.0^{20,25}]hexacosa-20,22,24-triene-3,7,15,26-tetrone, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Hargreaves, D. | | Deposit date: | 2016-11-16 | | Release date: | 2017-01-18 | | Last modified: | 2017-03-08 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors
Acs Med.Chem.Lett., 8, 2017
|
|
5GOG
 
 | | Lys29-linked di-ubiquitin | | Descriptor: | D-ubiquitin, Ubiquitin | | Authors: | Gao, S, Pan, M, Zheng, Y. | | Deposit date: | 2016-07-27 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
5GOK
 
 | | K11/K63-branched tri-Ubiquitin | | Descriptor: | D-ubiquitin, Ubiquitin | | Authors: | Gao, S, Pan, M, Zheng, Y, Liu, L. | | Deposit date: | 2016-07-27 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
5GOB
 
 | | Lys6-linked di-ubiquitin | | Descriptor: | D-ubiquitin, Ubiquitin | | Authors: | Gao, S, Pan, M, Zheng, Y, Liu, L. | | Deposit date: | 2016-07-26 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.153 Å) | | Cite: | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
5GOH
 
 | | Lys33-linked di-ubiquitin | | Descriptor: | D-ubiquitin, Ubiquitin | | Authors: | Gao, S, Pan, M, Zheng, Y, Liu, L. | | Deposit date: | 2016-07-27 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.946 Å) | | Cite: | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
5GO8
 
 | | Linear tetra-ubiquitin | | Descriptor: | D-ubiquitin, ubiquitin | | Authors: | Gao, S, Pan, M, Zheng, Y. | | Deposit date: | 2016-07-26 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
5GOD
 
 | | Lys27-linked di-ubiquitin | | Descriptor: | D-ubiquitin, MAGNESIUM ION, Ubiquitin | | Authors: | Gao, S, Pan, M, Zheng, Y. | | Deposit date: | 2016-07-26 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
5GOJ
 
 | | Lys63-linked di-ubiquitin | | Descriptor: | D-ubiquitin, Ubiquitin | | Authors: | Gao, S, Pan, M, Zheng, Y, Liu, L. | | Deposit date: | 2016-07-27 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
5GO7
 
 | | Linear tri-ubiquitin | | Descriptor: | D-ubiquitin, Ubiquitin | | Authors: | Gao, S, Pan, M, Zheng, Y. | | Deposit date: | 2016-07-26 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
5GOC
 
 | | Lys11-linked diubiquitin | | Descriptor: | D-ubiquitin, SODIUM ION, Ubiquitin | | Authors: | Gao, S, Pan, M, Zheng, Y. | | Deposit date: | 2016-07-26 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
5GOI
 
 | | Lys48-linked di-ubiquitin | | Descriptor: | D-ubiquitin, Ubiquitin | | Authors: | Gao, S, Pan, M, Zheng, Y, Liu, L. | | Deposit date: | 2016-07-27 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
4FMQ
 
 | |
6DE4
 
 | | Homo sapiens dihydrofolate reductase complexed with beta-NADPH and 3'-[(2R)-4-(2,4-diamino-6-ethylphenyl)but-3-yn-2-yl]-5'-methoxy-[1,1'-biphenyl]-4-carboxylic acid | | Descriptor: | 3'-[(2R)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl]-5'-methoxy[1,1'-biphenyl]-4-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hajian, B, Wright, D. | | Deposit date: | 2018-05-11 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Drugging the Folate Pathway in Mycobacterium tuberculosis: The Role of Multi-targeting Agents.
Cell Chem Biol, 26, 2019
|
|
6DDS
 
 | | Mycobacterium tuberculosis Dihydrofolate Reductase complexed with beta-NADPH and 4-[3-[3-[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]prop-2-ynyl]-5-methoxy-phenyl]benzoic acid | | Descriptor: | 4-[3-[3-[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]prop-2-ynyl]-5-methoxy-phenyl]benzoic acid, ACETATE ION, Dihydrofolate reductase, ... | | Authors: | Hajian, B, Wright, D, Scocchera, E. | | Deposit date: | 2018-05-10 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Drugging the Folate Pathway in Mycobacterium tuberculosis: The Role of Multi-targeting Agents.
Cell Chem Biol, 26, 2019
|
|
6DDW
 
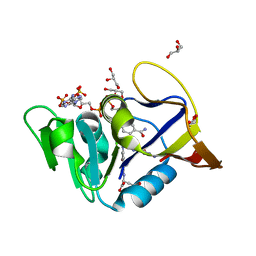 | | Mycobacterium tuberculosis Dihydrofolate Reductase complexed with beta-NADPH and N-(4-{[(2-amino-4-oxo-3,4-dihydropteridin-6-yl)methyl]amino}-2-hydroxybenzene-1-carbonyl)-L-glutamic acid | | Descriptor: | Dihydrofolate reductase, GLYCEROL, N-(4-{[(2-amino-4-oxo-3,4-dihydropteridin-6-yl)methyl]amino}-2-hydroxybenzene-1-carbonyl)-L-glutamic acid, ... | | Authors: | Hajian, B, Scocchera, E, Wright, D. | | Deposit date: | 2018-05-10 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Drugging the Folate Pathway in Mycobacterium tuberculosis: The Role of Multi-targeting Agents.
Cell Chem Biol, 26, 2019
|
|
5VH3
 
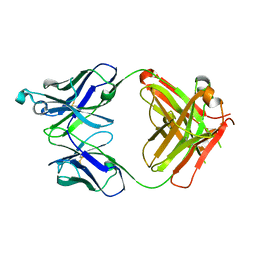 | | Crystal structure of Fab fragment of the anti-TNFa antibody infliximab in a C-centered orthorhombic crystal form | | Descriptor: | 1,2-ETHANEDIOL, Infliximab Fab Heavy Chain, Infliximab Fab Light Chain | | Authors: | Mayclin, S.J, Edwards, T.E, Lerch, T.F, Conlan, H, Sharpe, P. | | Deposit date: | 2017-04-12 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Infliximab crystal structures reveal insights into self-association.
MAbs, 9, 2017
|
|
5VH4
 
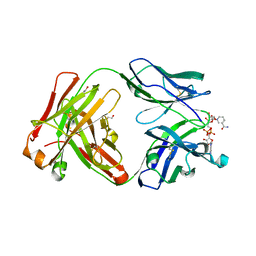 | | Crystal structure of Fab fragment of anti-TNFa antibody infliximab in an I-centered orthorhombic crystal form | | Descriptor: | 1,2-ETHANEDIOL, Infliximab Fab Heavy Chain, Infliximab Fab Light Chain, ... | | Authors: | Mayclin, S.J, Edwards, T.E, Lerch, T.F, Conlan, H, Sharpe, P. | | Deposit date: | 2017-04-12 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Infliximab crystal structures reveal insights into self-association.
MAbs, 9, 2017
|
|
2XRW
 
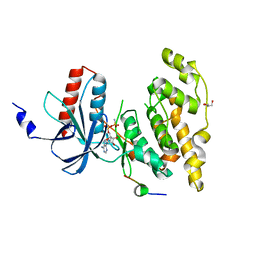 | | Linear binding motifs for JNK and for calcineurin antagonistically control the nuclear shuttling of NFAT4 | | Descriptor: | GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE 8, NUCLEAR FACTOR OF ACTIVATED T-CELLS, ... | | Authors: | Barkai, T, Toeoroe, I, Garai, A, Remenyi, A. | | Deposit date: | 2010-09-23 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Specificity of Linear Motifs that Bind to a Common Mitogen-Activated Protein Kinase Docking Groove.
Sci. Signal, 5, 2012
|
|
2XS0
 
 | | Linear binding motifs for JNK and for calcineurin antagonistically control the nuclear shuttling of NFAT4 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 8, NUCLEAR FACTOR OF ACTIVATED T-CELLS, CYTOPLASMIC 3, ... | | Authors: | Barkai, T, Toeoroe, I, Garai, A, Remenyi, A. | | Deposit date: | 2010-09-24 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Specificity of Linear Motifs that Bind to a Common Mitogen-Activated Protein Kinase Docking Groove.
Sci. Signal, 5, 2012
|
|
7QTZ
 
 | | Crystal structure of Iripin-1 serpin from tick Ixodes ricinus | | Descriptor: | MAGNESIUM ION, Putative salivary serpin | | Authors: | Kascakova, B, Kuta Smatanova, I, Chmelar, J, Prudnikova, T. | | Deposit date: | 2022-01-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Iripin-1, a new anti-inflammatory tick serpin, inhibits leukocyte recruitment in vivo while altering the levels of chemokines and adhesion molecules.
Front Immunol, 14, 2023
|
|
8G0M
 
 | | Structure of complex between TV6.6 and CD98hc ECD | | Descriptor: | 1,2-ETHANEDIOL, 4F2 cell-surface antigen heavy chain, TETRAETHYLENE GLYCOL, ... | | Authors: | Kariolis, M.S, Lexa, K, Liau, N.P.D, Srivastava, D, Tran, H, Wells, R.C. | | Deposit date: | 2023-01-31 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | CD98hc is a target for brain delivery of biotherapeutics.
Nat Commun, 14, 2023
|
|
