7C2M
 
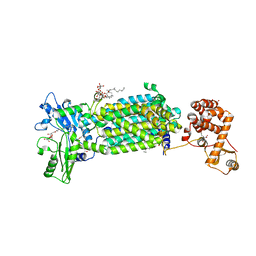 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with NITD-349 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Chimera of drug exporters of the RND superfamily-like protein and Endolysin, N-(4,4-dimethylcyclohexyl)-4,6-bis(fluoranyl)-1H-indole-2-carboxamide, ... | | Authors: | Zhang, B, Yang, X, Hu, T, Rao, Z. | | Deposit date: | 2020-05-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for the Inhibition of Mycobacterial MmpL3 by NITD-349 and SPIRO.
J.Mol.Biol., 432, 2020
|
|
7ENV
 
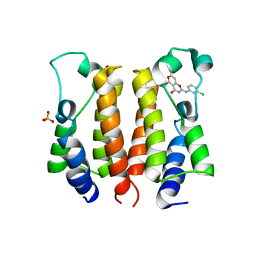 | | crystal structure of NS5 in complex with the N-terminal bromodomain of BRD2 (BRD2-BD1). | | Descriptor: | 7-chloranyl-2-[(3-chlorophenyl)amino]pyrano[3,4-e][1,3]oxazine-4,5-dione, Bromodomain-containing protein 2, SULFATE ION | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S. | | Deposit date: | 2021-04-19 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural investigation of a pyrano-1,3-oxazine derivative and the phenanthridinone core moiety against BRD2 bromodomains.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7EO5
 
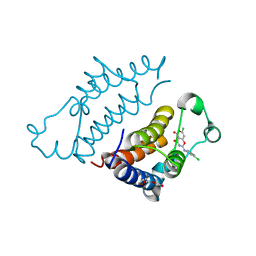 | | Crystal structure of pyrano 1,3, oxazine derivative in complex with the second bromodomain of BRD2 | | Descriptor: | 7-chloranyl-2-[(3-chlorophenyl)amino]pyrano[3,4-e][1,3]oxazine-4,5-dione, Bromodomain-containing protein 2, TRIETHYLENE GLYCOL | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of a pyrano-1,3-oxazine derivative and the phenanthridinone core moiety against BRD2 bromodomains.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7VRM
 
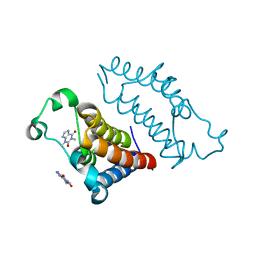 | | crystal structure of BRD2-BD2 in complex with purine derivative | | Descriptor: | Bromodomain-containing protein 2, THEOPHYLLINE | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-23 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7VRO
 
 | | crystal structure of BRD2-BD1 in complex with purine derivative | | Descriptor: | Bromodomain-containing protein 2, SULFATE ION, THEOBROMINE | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-23 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7VRQ
 
 | | crystal structure of BRD2-BD2 in complex with purine derivative | | Descriptor: | Bromodomain-containing protein 2, GLYCEROL, THEOBROMINE | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-23 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7VSF
 
 | | crystal structure of BRD2-BD1 in complex with purine derivative | | Descriptor: | 3-methyl-7-propyl-purine-2,6-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bromodomain-containing protein 2, ... | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-26 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7VS0
 
 | | crystal structure of BRD2-BD2 in complex with purine derivative | | Descriptor: | Bromodomain-containing protein 2, Doxofylline, GLYCEROL | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7VRK
 
 | | crystal structure of BRD2-BD1 in complex with purine derivative | | Descriptor: | Bromodomain-containing protein 2, SULFATE ION, THEOPHYLLINE | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-23 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7VRH
 
 | | crystal structure of BRD2-BD1 in complex with guanosine analog | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 9-HYROXYETHOXYMETHYLGUANINE, Bromodomain-containing protein 2, ... | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-22 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7VS1
 
 | | crystal structure of BRD2-BD2 in complex with purine derivative | | Descriptor: | 3-methyl-7-propyl-purine-2,6-dione, Bromodomain-containing protein 2, GLYCEROL | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7VRI
 
 | | crystal structure of BRD2-BD2 in complex with guanosine analog | | Descriptor: | 9-HYROXYETHOXYMETHYLGUANINE, Bromodomain-containing protein 2 | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-23 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7VRZ
 
 | | crystal structure of BRD2-BD1 in complex with purine derivative | | Descriptor: | Bromodomain-containing protein 2, Doxofylline, SULFATE ION | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7WNX
 
 | | Cryo-EM structure of Mycobacterium smegmatis MmpL3 complexed with ST004 in lipid nanodiscs | | Descriptor: | N-[2-(2-adamantylamino)ethyl]-1-[2,4-bis(fluoranyl)phenyl]-5-(4-chlorophenyl)-4-methyl-pyrazole-3-carboxamide, Trehalose monomycolate exporter MmpL3 | | Authors: | Zhang, B, Hu, T, Yang, X, Liu, F, Rao, Z. | | Deposit date: | 2022-01-20 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structure-based design of anti-mycobacterial drug leads that target the mycolic acid transporter MmpL3.
Structure, 30, 2022
|
|
7E36
 
 | | A [6+4]-cycloaddition adduct is the biosynthetic intermediate in streptoseomycin biosynthesis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alkanesulfonate monooxygenase SsuD/methylene tetrahydromethanopterin reductase-like flavin-dependent oxidoreductase (Luciferase family), ... | | Authors: | Zhang, B, Ge, H.M. | | Deposit date: | 2021-02-08 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A [6+4]-cycloaddition adduct is the biosynthetic intermediate in streptoseomycin biosynthesis.
Nat Commun, 12, 2021
|
|
7FE6
 
 | |
7FE0
 
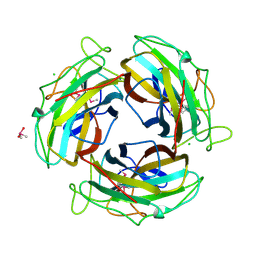 | |
7C2N
 
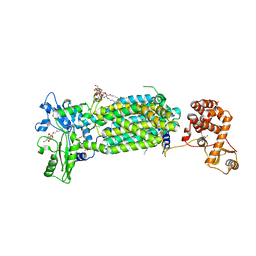 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with SPIRO | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 1'-(2,3-dihydro-1,4-benzodioxin-6-ylmethyl)spiro[6,7-dihydrothieno[3,2-c]pyran-4,4'-piperidine], Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Yang, X, Hu, T, Rao, Z. | | Deposit date: | 2020-05-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural Basis for the Inhibition of Mycobacterial MmpL3 by NITD-349 and SPIRO.
J.Mol.Biol., 432, 2020
|
|
7C5E
 
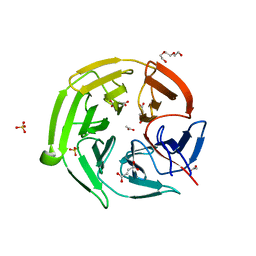 | | Crystal structure of Keap1 in complex with fumarate (FUM) | | Descriptor: | ACETATE ION, FUMARIC ACID, Kelch-like ECH-associated protein 1, ... | | Authors: | Padmanabhan, B, Unni, S, Deshmukh, P. | | Deposit date: | 2020-05-19 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the multiple binding modes of Dimethyl Fumarate (DMF) and its analogs to the Kelch domain of Keap1.
Febs J., 288, 2021
|
|
7WIU
 
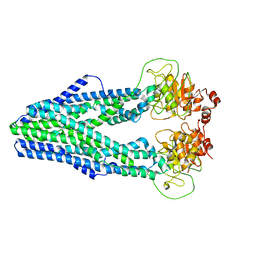 | | Cryo-EM structure of Mycobacterium tuberculosis irtAB in inward-facing state | | Descriptor: | Mycobactin import ATP-binding/permease protein IrtA, Mycobactin import ATP-binding/permease protein IrtB | | Authors: | Zhang, B, Sun, S, Yang, H, Rao, Z. | | Deposit date: | 2022-01-05 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Cryo-EM structures for the Mycobacterium tuberculosis iron-loaded siderophore transporter IrtAB.
Protein Cell, 14, 2023
|
|
7WIX
 
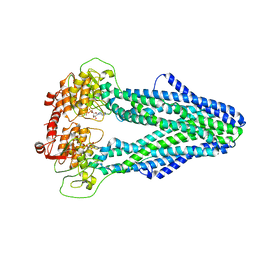 | | Cryo-EM structure of Mycobacterium tuberculosis irtAB in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Mycobactin import ATP-binding/permease protein IrtA, ... | | Authors: | Zhang, B, Sun, S, Yang, H, Rao, Z. | | Deposit date: | 2022-01-05 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Cryo-EM structures for the Mycobacterium tuberculosis iron-loaded siderophore transporter IrtAB.
Protein Cell, 14, 2023
|
|
7WIV
 
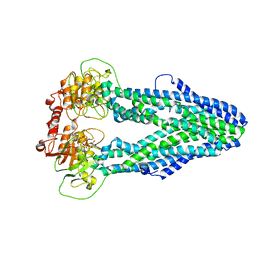 | | Cryo-EM structure of Mycobacterium tuberculosis irtAB in complex with an AMP-PNP | | Descriptor: | Mycobactin import ATP-binding/permease protein IrtA, Mycobactin import ATP-binding/permease protein IrtB, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Zhang, B, Sun, S, Yang, H, Rao, Z. | | Deposit date: | 2022-01-05 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Cryo-EM structures for the Mycobacterium tuberculosis iron-loaded siderophore transporter IrtAB.
Protein Cell, 14, 2023
|
|
7WIW
 
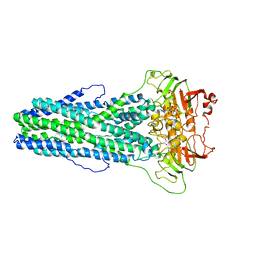 | | Cryo-EM structure of Mycobacterium tuberculosis irtAB complexed with ATP in an occluded conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mycobactin import ATP-binding/permease protein IrtA, ... | | Authors: | Zhang, B, Sun, S, Yang, H, Rao, Z. | | Deposit date: | 2022-01-05 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structures for the Mycobacterium tuberculosis iron-loaded siderophore transporter IrtAB.
Protein Cell, 14, 2023
|
|
7FE5
 
 | |
7C60
 
 | | Crystal structure of Keap1 in complex with monoethyl fumarate (MEF) | | Descriptor: | (~{Z})-4-ethoxy-4-oxidanylidene-but-2-enoic acid, ACETATE ION, Kelch-like ECH-associated protein 1, ... | | Authors: | Padmanabhan, B, Unni, S, Deshmukh, P, Krishnappa, G. | | Deposit date: | 2020-05-21 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into the multiple binding modes of Dimethyl Fumarate (DMF) and its analogs to the Kelch domain of Keap1.
Febs J., 288, 2021
|
|
