8W53
 
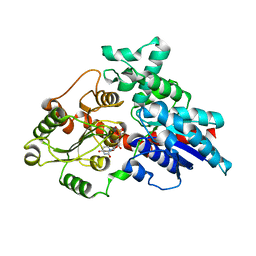 | | Crystal structure of LbUGT in complex with UDP | | Descriptor: | GLYCEROL, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Hu, D, Wang, G.Q. | | Deposit date: | 2023-08-25 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.42816162 Å) | | Cite: | Functional and structural dissection of glycosyltransferases underlying the glycodiversity of wolfberry-derived bioactive ingredients lycibarbarspermidines.
Nat Commun, 15, 2024
|
|
5VEB
 
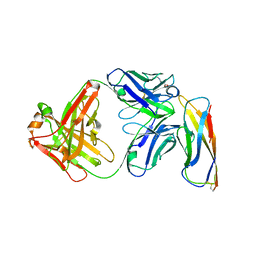 | | Crystal structure of a Fab binding to extracellular domain 5 of Cadherin-6 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Cadherin-6, anti-CDH6 Fab heavy chain, ... | | Authors: | Zhu, X, Bialucha, C.U, London, A, Clark, K, Hu, T. | | Deposit date: | 2017-04-04 | | Release date: | 2017-06-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Discovery and Optimization of HKT288, a Cadherin-6-Targeting ADC for the Treatment of Ovarian and Renal Cancers.
Cancer Discov, 7, 2017
|
|
5UZK
 
 | | Crystal Structure of PKA bound to an pyrrolo pyridine inhibitor | | Descriptor: | 2-{3-[3-(piperidin-4-yl)propoxy]phenyl}-N-[4-(1H-pyrrolo[2,3-b]pyridin-3-yl)-1,3-thiazol-2-yl]acetamide, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Jacobs, M.D, Brown, K. | | Deposit date: | 2017-02-26 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ROCK inhibitors 3: Design, synthesis and structure-activity relationships of 7-azaindole-based Rho kinase (ROCK) inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
5EHR
 
 | |
5F7E
 
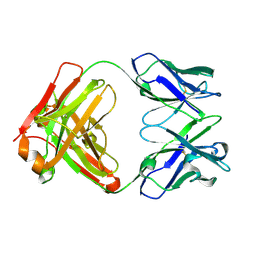 | | Crystal structure of germ-line precursor of 3BNC60 Fab | | Descriptor: | Fab heavy chain, Fab light chain | | Authors: | Sievers, S.A, Scharf, L, Jiang, S, Bjorkman, P.J. | | Deposit date: | 2015-12-08 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for germline antibody recognition of HIV-1 immunogens.
Elife, 5, 2016
|
|
5UZJ
 
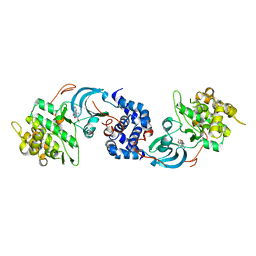 | | Crystal Structure of ROCK1 bound to an aminopyridine inhibitor | | Descriptor: | N-[4-(2-aminopyridin-4-yl)-1,3-thiazol-2-yl]-2-(3-methoxyphenyl)acetamide, Rho-associated protein kinase 1 | | Authors: | Jacobs, M.D. | | Deposit date: | 2017-02-26 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | ROCK inhibitors 3: Design, synthesis and structure-activity relationships of 7-azaindole-based Rho kinase (ROCK) inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
7U0K
 
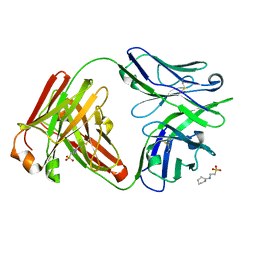 | | IOMA class antibody Fab ACS124 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, IOMA Class antibody ACS124 Heavy chain, IOMA Class antibody ACS124 Light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.
Nat Commun, 13, 2022
|
|
7U04
 
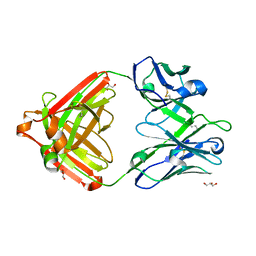 | | IOMA class antibody ACS101 | | Descriptor: | GLYCEROL, IOMA class antibody ACS101 heavy chain, IOMA class antibody ACS101 light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-02-17 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.
Nat Commun, 13, 2022
|
|
7YFN
 
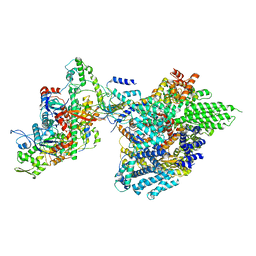 | | Core module of the NuA4 complex in S. cerevisiae | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ARP4 isoform 1, Actin, ... | | Authors: | Ji, L.T, Zhao, L.X, Xu, K, Gao, H.H, Zhou, Y, Kornberg, R.D, Zhang, H.Q. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the NuA4 histone acetyltransferase complex.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7EN0
 
 | | Structure and Activity of SLAC1 Channels for Stomatal Signaling in Leaves | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, SLow Anion Channel 1, SPHINGOSINE | | Authors: | Deng, Y, Kashtoh, H, Wang, Q, Zhen, G, Li, Q, Tang, L, Gao, H, Zhang, C, Qin, L, Su, M, Li, F, Huang, X, Wang, Y, Xie, Q, Clarke, O.B, Hendrickson, W.A, Chen, Y. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure and activity of SLAC1 channels for stomatal signaling in leaves.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7YFP
 
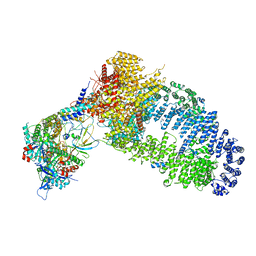 | | The NuA4 histone acetyltransferase complex from S. cerevisiae | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ARP4 isoform 1, Actin, ... | | Authors: | Ji, L.T, Zhao, L.X, Xu, K, Gao, H.H, Zhou, Y, Kornberg, R.D, Zhang, H.Q. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the NuA4 histone acetyltransferase complex.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4P9M
 
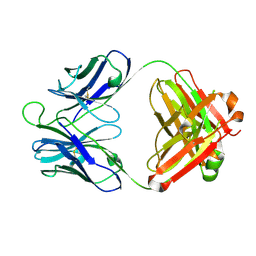 | | Crystal structure of 8ANC195 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8ANC195 Fab heavy chain, 8ANC195 light chain | | Authors: | Scharf, L, Bjorkman, P.J. | | Deposit date: | 2014-04-04 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Antibody 8ANC195 Reveals a Site of Broad Vulnerability on the HIV-1 Envelope Spike.
Cell Rep, 7, 2014
|
|
4P9H
 
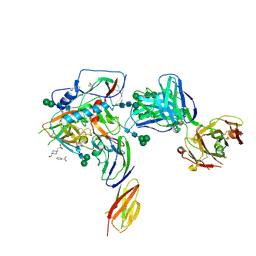 | |
3U7Y
 
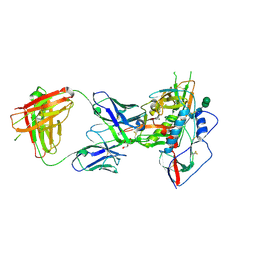 | |
3U7W
 
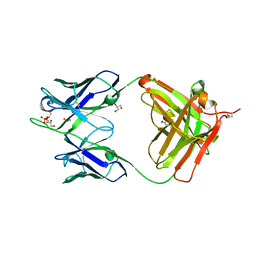 | | Crystal structure of NIH45-46 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Heavy chain, ... | | Authors: | Diskin, R, Bjorkman, P.J. | | Deposit date: | 2011-10-14 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Increasing the Potency and Breadth of an HIV Antibody by Using Structure-Based Rational Design.
Science, 334, 2011
|
|
4JDT
 
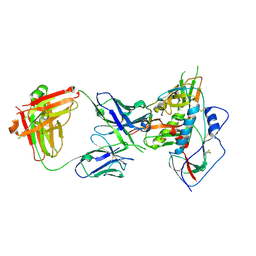 | | Crystal structure of chimeric germ-line precursor of NIH45-46 Fab in complex with gp120 of 93TH057 HIV-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, Fab light chain, ... | | Authors: | Scharf, L, Diskin, R, Bjorkman, P.J. | | Deposit date: | 2013-02-25 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Structural basis for HIV-1 gp120 recognition by a germ-line version of a broadly neutralizing antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JKP
 
 | |
4JDV
 
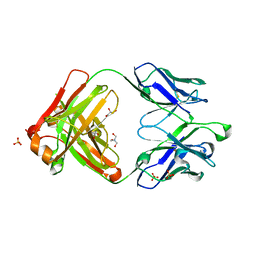 | | Crystal structure of germ-line precursor of NIH45-46 Fab | | Descriptor: | Fab heavy chain, Fab light chain, GLYCEROL, ... | | Authors: | Diskin, R, Scharf, L, Bjorkman, P.J. | | Deposit date: | 2013-02-25 | | Release date: | 2013-03-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for HIV-1 gp120 recognition by a germ-line version of a broadly neutralizing antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6B2Q
 
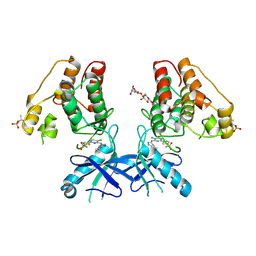 | | Dual Inhibition of the Essential Protein Kinases A and B in Mycobacterium tuberculosis | | Descriptor: | 3-methyl-1-(2-methylpropyl)butyl 4-O-beta-L-gulopyranosyl-beta-D-glucopyranoside, 5-{5-chloro-4-[(5-cyclopropyl-1H-pyrazol-3-yl)amino]pyrimidin-2-yl}thiophene-2-sulfonamide, Serine/threonine-protein kinase PknA | | Authors: | Zuccola, H.J. | | Deposit date: | 2017-09-20 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Mtb PKNA/PKNB Dual Inhibition Provides Selectivity Advantages for Inhibitor Design To Minimize Host Kinase Interactions.
ACS Med Chem Lett, 8, 2017
|
|
6B2P
 
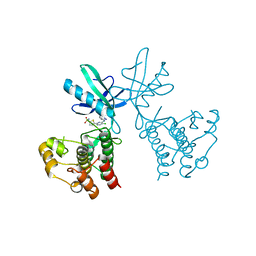 | | Dual Inhibition of the Essential Protein Kinases A and B in Mycobacterium tuberculosis | | Descriptor: | 5-{5-chloro-4-[(5-cyclopropyl-1H-pyrazol-3-yl)amino]pyrimidin-2-yl}thiophene-2-sulfonamide, Serine/threonine-protein kinase PknB | | Authors: | Zuccola, H.J. | | Deposit date: | 2017-09-20 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Mtb PKNA/PKNB Dual Inhibition Provides Selectivity Advantages for Inhibitor Design To Minimize Host Kinase Interactions.
ACS Med Chem Lett, 8, 2017
|
|
7CBL
 
 | | Cryo-EM structure of the flagellar LP ring from Salmonella | | Descriptor: | Flagellar L-ring protein, Flagellar P-ring protein, OCTANOIC ACID (CAPRYLIC ACID) | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-06-12 | | Release date: | 2021-04-28 | | Last modified: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CGB
 
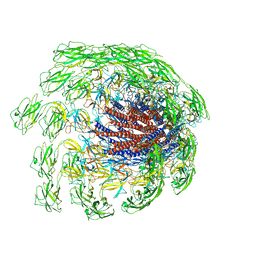 | | Cryo-EM structure of the flagellar hook from Salmonella | | Descriptor: | Flagellar hook protein FlgE | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-07-01 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CG7
 
 | | Cryo-EM structure of the flagellar MS ring with C34 symmetry from Salmonella | | Descriptor: | Flagellar M-ring protein | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-06-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CGO
 
 | | Cryo-EM structure of the flagellar motor-hook complex from Salmonella | | Descriptor: | Flagellar L-ring protein, Flagellar M-ring protein, Flagellar MS ring L1, ... | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-07-01 | | Release date: | 2021-04-28 | | Last modified: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CG4
 
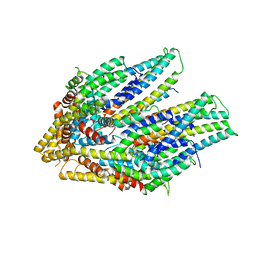 | | Cryo-EM structure of the flagellar export apparatus with FliE from Salmonella | | Descriptor: | Flagellar biosynthetic protein FliP, Flagellar hook-basal body complex protein FliE | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-06-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
