2AXK
 
 | | Solution structure of discrepin, a scorpion venom toxin blocking K+ channels. | | Descriptor: | discrepin | | Authors: | Prochnicka-Chalufour, A, Corzo, G, Satake, H, Martin-Eauclaire, M.-F, Murgia, A.R, Prestipino, G, D'Suze, G, Possani, L.D, Delepierre, M. | | Deposit date: | 2005-09-05 | | Release date: | 2006-06-20 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Solution structure of discrepin, a new K+-channel blocking peptide from the alpha-KTx15 subfamily.
Biochemistry, 45, 2006
|
|
4Q6U
 
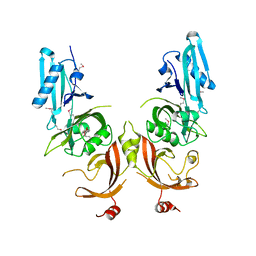 | |
4YPI
 
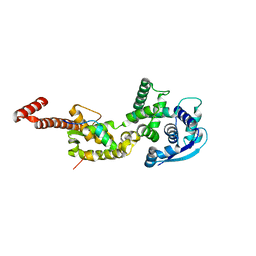 | | Structure of Ebola virus nucleoprotein N-terminal fragment bound to a peptide derived from Ebola VP35 | | Descriptor: | Nucleoprotein, Polymerase cofactor VP35 | | Authors: | Leung, D.W, Borek, D.M, Binning, J.M, Otwinowski, Z, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-03-13 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | An Intrinsically Disordered Peptide from Ebola Virus VP35 Controls Viral RNA Synthesis by Modulating Nucleoprotein-RNA Interactions.
Cell Rep, 11, 2015
|
|
3R7C
 
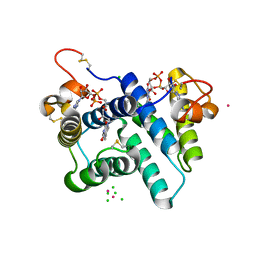 | | The structure of a hexahestidine-tagged form of augmenter of liver regeneration reveals a novel Cd(2)Cl(4)O(6) cluster that aids in crystal packing | | Descriptor: | CADMIUM ION, CHLORIDE ION, FAD-linked sulfhydryl oxidase ALR, ... | | Authors: | Florence, Q.J, Wu, C.-K, Swindell II, J.T, Wang, B.C, Rose, J.P. | | Deposit date: | 2011-03-22 | | Release date: | 2012-03-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of a hexahestidine-tagged form of augmenter of liver regeneration reveals a novel Cd(2)Cl(4)O(6) cluster that aids in crystal packing
To be Published
|
|
5AJB
 
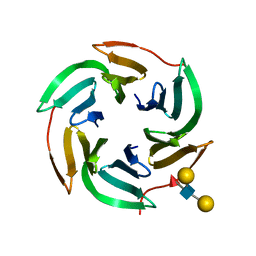 | | X-ray structure of the RSL lectin in complex with Lewis X tetrasaccahride | | Descriptor: | LECTIN, alpha-L-fucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Topin, J, Arnaud, J, Varrot, A, Imberty, A. | | Deposit date: | 2015-02-20 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Hidden Conformation of Lewis X, a Human Histo-Blood Group Antigen, is a Determinant for Recognition by Pathogen Lectins
Acs Chem.Biol., 11, 2016
|
|
5AJC
 
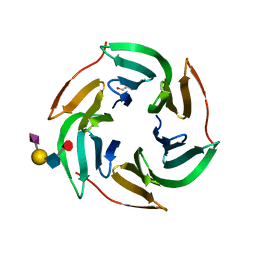 | | X-ray structure of RSL lectin in complex with sialyl lewis X tetrasaccharide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose, PUTATIVE FUCOSE-BINDING LECTIN PROTEIN, ... | | Authors: | Topin, J, Arnaud, J, Varrot, A, Imberty, A. | | Deposit date: | 2015-02-20 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Hidden Conformation of Lewis X, a Human Histo-Blood Group Antigen, is a Determinant for Recognition by Pathogen Lectins
Acs Chem.Biol., 11, 2016
|
|
4I3F
 
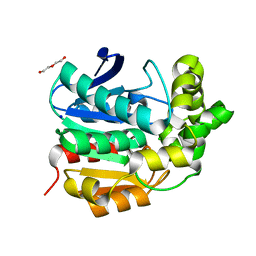 | | Crystal structure of serine hydrolase CCSP0084 from the polyaromatic hydrocarbon (PAH)-degrading bacterium Cycloclasticus zankles | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Stogios, P.J, Xu, X, Dong, A, Cui, H, Alcaide, M, Tornes, J, Gertler, C, Yakimov, M.M, Golyshin, P.N, Ferrer, M, Savchenko, A. | | Deposit date: | 2012-11-26 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Single residues dictate the co-evolution of dual esterases: MCP hydrolases from the alpha / beta hydrolase family.
Biochem.J., 454, 2013
|
|
8A9U
 
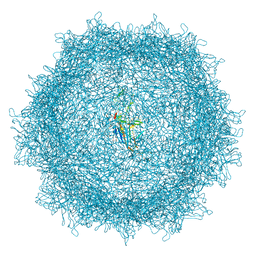 | | Full AAV3B-VP1KO virion | | Descriptor: | Capsid protein VP1 | | Authors: | Arriaga, I, Abrescia, N.G.A. | | Deposit date: | 2022-06-29 | | Release date: | 2022-09-21 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cellular and Structural Characterization of VP1 and VP2 Knockout Mutants of AAV3B Serotype and Implications for AAV Manufacturing.
Hum Gene Ther, 33, 2022
|
|
2ANJ
 
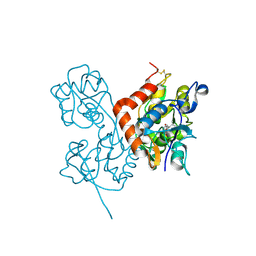 | | Crystal Structure of the Glur2 Ligand Binding Core (S1S2J-Y450W) Mutant in Complex With the Partial Agonist Kainic Acid at 2.1 A Resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2 | | Authors: | Holm, M.M, Naur, P, Vestergaard, B, Geballe, M.T, Gajhede, M, Kastrup, J.S, Traynelis, S.F, Egebjerg, J. | | Deposit date: | 2005-08-11 | | Release date: | 2005-08-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Binding Site Tyrosine Shapes Desensitization Kinetics and Agonist Potency at GluR2: a mutagenic, kinetic, and crystallographic study
J.Biol.Chem., 280, 2005
|
|
6SZA
 
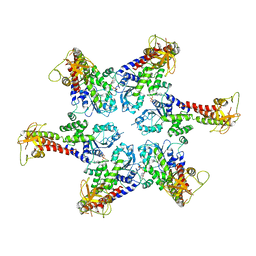 | |
6SZB
 
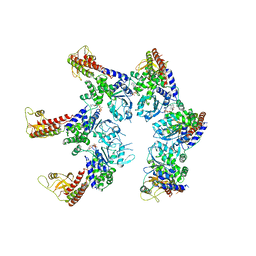 | |
7ZVB
 
 | | Crystal Structure of the mature form of the glutamic-class prolyl-endopeptidase neprosin at 2.35 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-terminal peptidase, TRIETHYLENE GLYCOL, ... | | Authors: | Del Amo-Maestro, L, Eckhard, U, Rodriguez-Banqueri, A, Mendes, S.R, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular and in vivo studies of a glutamate-class prolyl-endopeptidase for coeliac disease therapy.
Nat Commun, 13, 2022
|
|
7ZVC
 
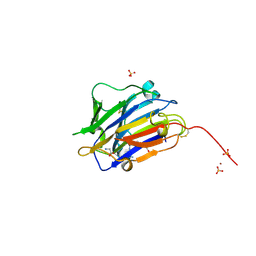 | | Second crystal form of the mature glutamic-class prolyl-endopeptidase neprosin at 1.85 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-terminal peptidase, GLY-GLY-GLY-GLY, ... | | Authors: | Rodriguez-Banqueri, A, Eckhard, U, Del Amo-Maestro, L, Mendes, S.R, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular and in vivo studies of a glutamate-class prolyl-endopeptidase for coeliac disease therapy.
Nat Commun, 13, 2022
|
|
7ZVA
 
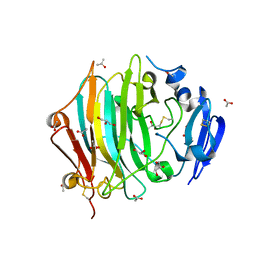 | | Crystal Structure of the native zymogen form of the glutamic-class prolyl-endopeptidase neprosin at 1.80 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, C-terminal peptidase, ... | | Authors: | Del Amo-Maestro, L, Eckhard, U, Rodriguez-Banqueri, A, Mendes, S.R, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular and in vivo studies of a glutamate-class prolyl-endopeptidase for coeliac disease therapy.
Nat Commun, 13, 2022
|
|
7ZU8
 
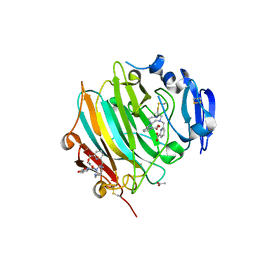 | | Crystal Structure of the zymogen form of the glutamic-class prolyl-endopeptidase neprosin at 2.05 A resolution in presence of the crystallophore Lu-Xo4. | | Descriptor: | 12-oxidanyl-9,11$l^{3}-dioxa-1$l^{4},19$l^{4},22,27$l^{4},28$l^{4}-pentaza-10$l^{6}-lutetaoctacyclo[17.5.2.1^{3,7}.1^{10,13}.0^{1,10}.0^{10,19}.0^{10,28}.0^{17,27}]octacosa-3,5,7(28),11,13,15,17(27)-heptaen-8-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Del Amo-Maestro, L, Eckhard, U, Rodriguez-Banqueri, A, Mendes, S.R, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2022-05-11 | | Release date: | 2022-08-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular and in vivo studies of a glutamate-class prolyl-endopeptidase for coeliac disease therapy.
Nat Commun, 13, 2022
|
|
4BMV
 
 | | Short-chain dehydrogenase from Sphingobium yanoikuyae in complex with NADPH | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SHORT-CHAIN DEHYDROGENASE | | Authors: | Man, H, Kedziora, K, Lavandera-Garcia, I, Gotor-Fernandez, V, Grogan, G. | | Deposit date: | 2013-05-10 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Alcohol Dehydrogenases from Ralstonia and Sphingobium Spp. Reveal the Molecular Basis for Their Recognition of 'Bulky-Bulky' Ketones
Top.Catal., 57, 2014
|
|
4BMS
 
 | | Short chain alcohol dehydrogenase from Ralstonia sp. DSM 6428 in complex with NADPH | | Descriptor: | ALCLOHOL DEHYDROGENASE/SHORT-CHAIN DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Man, H, Kulig, J, Rother, D, Grogan, G. | | Deposit date: | 2013-05-10 | | Release date: | 2014-03-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structures of Alcohol Dehydrogenases from Ralstonia and Sphingobium Spp. Reveal the Molecular Basis for Their Recognition of 'Bulky-Bulky' Ketones
Top.Catal., 57, 2014
|
|
4BMN
 
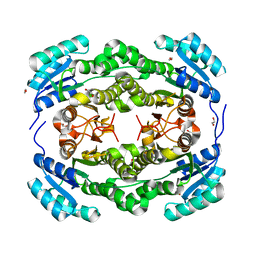 | | apo structure of short-chain alcohol dehydrogenase from Ralstonia sp. DSM 6428 | | Descriptor: | 1,2-ETHANEDIOL, ALCLOHOL DEHYDROGENASE/SHORT-CHAIN DEHYDROGENASE, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Man, H, Kulig, J, Rother, D, Grogan, G. | | Deposit date: | 2013-05-10 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of Alcohol Dehydrogenases from Ralstonia and Sphingobium Spp. Reveal the Molecular Basis for Their Recognition of 'Bulky-Bulky' Ketones
Top.Catal., 57, 2014
|
|
5MGQ
 
 | |
