3UY9
 
 | | Bovine trypsin variant X(tripleGlu217Phe227) in complex with small molecule inhibitor | | Descriptor: | BENZAMIDINE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-12-06 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
3V12
 
 | | Bovine trypsin variant X(tripleGlu217Phe227) in complex with small molecule inhibitor | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-12-09 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
3UNQ
 
 | | Bovine trypsin variant X(triplePhe227) in complex with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, BENZAMIDINE, CALCIUM ION, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-11-16 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
3UUZ
 
 | | Bovine trypsin variant X(triplePhe227) in complex with small molecule inhibitor | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-11-29 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
3UNS
 
 | | Bovine trypsin variant X(triplePhe227) in complex with small molecule inhibitor | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-11-16 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
3UOP
 
 | | Bovine trypsin variant X(triplePhe227) in complex with small molecule inhibitor | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cationic trypsin, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-11-17 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
3V13
 
 | | Bovine trypsin variant X(tripleGlu217Phe227) in complex with small molecule inhibitor | | Descriptor: | 3-(3-carbamimidoylphenyl)-N-(2'-sulfamoylbiphenyl-4-yl)-1,2-oxazole-4-carboxamide, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-12-09 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
4OMD
 
 | |
5L7T
 
 | | 17beta-hydroxysteroid dehydrogenase 14 variant T205 in complex with a non-steroidal inhibitor. | | Descriptor: | (4-fluoranyl-3-oxidanyl-phenyl)-[6-(3-methyl-4-oxidanyl-phenyl)pyridin-2-yl]methanone, 17-beta-hydroxysteroid dehydrogenase 14, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Bertoletti, N, Braun, F, Marchais-Oberwinkler, S, Heine, A, Klebe, G. | | Deposit date: | 2016-06-03 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | First Structure-Activity Relationship of 17 beta-Hydroxysteroid Dehydrogenase Type 14 Nonsteroidal Inhibitors and Crystal Structures in Complex with the Enzyme.
J. Med. Chem., 59, 2016
|
|
5L7Y
 
 | | 17beta-hydroxysteroid dehydrogenase 14 variant T205 in complex with a non-steroidal inhibitor. | | Descriptor: | (4-fluoranyl-3-oxidanyl-phenyl)-[6-(2-fluoranyl-3-oxidanyl-phenyl)pyridin-2-yl]methanone, 17-beta-hydroxysteroid dehydrogenase 14, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Bertoletti, N, Braun, F, Marchais-Oberwinkler, S, Heine, A, Klebe, G. | | Deposit date: | 2016-06-04 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.912 Å) | | Cite: | First Structure-Activity Relationship of 17 beta-Hydroxysteroid Dehydrogenase Type 14 Nonsteroidal Inhibitors and Crystal Structures in Complex with the Enzyme.
J. Med. Chem., 59, 2016
|
|
4OMC
 
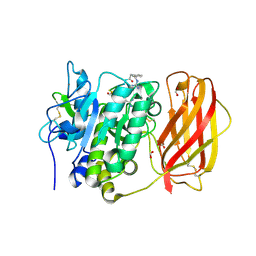 | |
5L7W
 
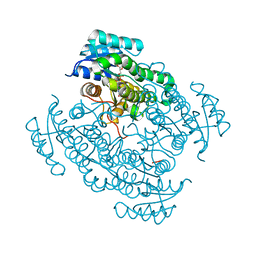 | | 17beta-hydroxysteroid dehydrogenase 14 variant T205 in complex with a non-steroidal inhibitor. | | Descriptor: | 17-beta-hydroxysteroid dehydrogenase 14, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Bertoletti, N, Braun, F, Marchais-Oberwinkler, S, Heine, A, Klebe, G. | | Deposit date: | 2016-06-04 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | First Structure-Activity Relationship of 17 beta-Hydroxysteroid Dehydrogenase Type 14 Nonsteroidal Inhibitors and Crystal Structures in Complex with the Enzyme.
J. Med. Chem., 59, 2016
|
|
7THS
 
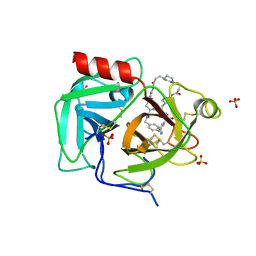 | | Macrocyclic plasmin inhibitor | | Descriptor: | (2S)-butane-1,2-diol, (6S,9R,20R,23S)-N-{[4-(aminomethyl)phenyl]methyl}-20-[(benzenesulfonyl)amino]-3,13,21-trioxo-2,6,9,14,22-pentaazatetracyclo[23.2.2.2~6,9~.2~15,18~]tritriaconta-1(27),15,17,25,28,30-hexaene-23-carboxamide, Plasminogen, ... | | Authors: | Guojie, W. | | Deposit date: | 2022-01-12 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and Structural Characterization of Macrocyclic Plasmin Inhibitors.
Chemmedchem, 18, 2023
|
|
6EQX
 
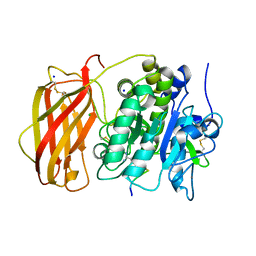 | |
6EQW
 
 | |
6EQV
 
 | |
7VLG
 
 | | Crystal structure of Zika NS2B-NS3 protease with compound MI2201 | | Descriptor: | 1-[(5~{R},8~{R},15~{S},18~{S})-15,18-bis(4-azanylbutyl)-4,7,14,17,20-pentakis(oxidanylidene)-5-propan-2-yl-3,6,13,16,19-pentazabicyclo[20.3.1]hexacosa-1(25),22(26),23-trien-8-yl]guanidine, NS3 protease, SULFATE ION, ... | | Authors: | Quek, J.P. | | Deposit date: | 2021-10-02 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | Structure-Based Optimization and Characterization of Macrocyclic Zika Virus NS2B-NS3 Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
7VLH
 
 | | Crystal structure of Zika NS2B-NS3 protease with compound MI2219 | | Descriptor: | 1-[(3~{S},6~{R},18~{R})-3,6-bis(4-azanylbutyl)-2,5,8,11,14,17-hexakis(oxidanylidene)-1,4,7,10,13,16-hexazacyclodocos-18-yl]guanidine, NS3 protease, SULFATE ION, ... | | Authors: | Quek, J.P. | | Deposit date: | 2021-10-02 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.621 Å) | | Cite: | Structure-Based Optimization and Characterization of Macrocyclic Zika Virus NS2B-NS3 Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
3RLW
 
 | | Human Thrombin in complex with MI328 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hirudin variant-2, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-04-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Ligand binding stepwise disrupts water network in thrombin: enthalpic and entropic changes reveal classical hydrophobic effect
J.Med.Chem., 55, 2012
|
|
3RMO
 
 | | Human Thrombin in complex with MI004 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hirudin variant-2, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-04-21 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Ligand binding stepwise disrupts water network in thrombin: enthalpic and entropic changes reveal classical hydrophobic effect
J.Med.Chem., 55, 2012
|
|
7UAH
 
 | | Macrocyclic plasmin inhibitor | | Descriptor: | (2~{R})-butane-1,2-diol, (6S,9R,19S,22R)-N-{[4-(aminomethyl)phenyl]methyl}-22-[(3-chlorobenzene-1-sulfonyl)amino]-3,12,21-trioxo-2,6,9,13,20-pentaazatetracyclo[22.2.2.2~6,9~.2~14,17~]dotriaconta-1(26),14,16,24,27,29-hexaene-19-carboxamide, Plasminogen, ... | | Authors: | Guojie, W. | | Deposit date: | 2022-03-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Synthesis and Structural Characterization of Macrocyclic Plasmin Inhibitors.
Chemmedchem, 18, 2023
|
|
3RMM
 
 | | Human Thrombin in complex with MI332 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hirudin variant-2, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-04-21 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Ligand binding stepwise disrupts water network in thrombin: enthalpic and entropic changes reveal classical hydrophobic effect
J.Med.Chem., 55, 2012
|
|
3RM2
 
 | | Human Thrombin in complex with MI003 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hirudin variant-2, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-04-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Ligand binding stepwise disrupts water network in thrombin: enthalpic and entropic changes reveal classical hydrophobic effect
J.Med.Chem., 55, 2012
|
|
3RM0
 
 | | Human Thrombin in complex with MI354 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hirudin variant-2, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-04-20 | | Release date: | 2012-04-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Ligand binding stepwise disrupts water network in thrombin: enthalpic and entropic changes reveal classical hydrophobic effect
J.Med.Chem., 55, 2012
|
|
7VLI
 
 | | Crystal structure of Zika NS2B-NS3 protease with compound MI2220 | | Descriptor: | 1-[(3~{S},6~{S},19~{R})-3,6-bis(4-azanylbutyl)-2,5,8,12,15,18-hexakis(oxidanylidene)-1,4,7,11,14,17-hexazacyclotricos-19-yl]guanidine, NS3 protease, SULFATE ION, ... | | Authors: | Quek, J.P. | | Deposit date: | 2021-10-02 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | Structure-Based Optimization and Characterization of Macrocyclic Zika Virus NS2B-NS3 Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
