1JRU
 
 | | NMR STRUCTURE OF THE UBX DOMAIN FROM P47 (ENERGY MINIMISED AVERAGE) | | Descriptor: | p47 protein | | Authors: | Yuan, X.M, Shaw, A, Zhang, X.D, Kondo, H, Lally, J, Freemont, P.S, Matthews, S.J. | | Deposit date: | 2001-08-15 | | Release date: | 2001-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and interaction surface of the C-terminal domain from p47: a major p97-cofactor involved in SNARE disassembly.
J.Mol.Biol., 311, 2001
|
|
3B7B
 
 | | EuHMT1 (Glp) Ankyrin Repeat Domain (Structure 1) | | Descriptor: | Euchromatic histone-lysine N-methyltransferase 1, SULFATE ION | | Authors: | Collins, R.E, Horton, J.R, Cheng, X. | | Deposit date: | 2007-10-30 | | Release date: | 2008-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules
Nat.Struct.Mol.Biol., 15, 2008
|
|
5IME
 
 | | Crystal structure of P21-activated kinase 1 (PAK1) in complex with compound 9 | | Descriptor: | 8-(3-aminopropyl)-6-[2-chloro-4-(3-methyl-2-oxopyrazin-1(2H)-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7(8H)-one, Serine/threonine-protein kinase PAK 1 | | Authors: | Li, D, Wang, W. | | Deposit date: | 2016-03-06 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.217 Å) | | Cite: | Chemically Diverse Group I p21-Activated Kinase (PAK) Inhibitors Impart Acute Cardiovascular Toxicity with a Narrow Therapeutic Window.
J.Med.Chem., 59, 2016
|
|
3B95
 
 | | EuHMT1 (Glp) Ankyrin Repeat Domain (Structure 2) | | Descriptor: | Euchromatic histone-lysine N-methyltransferase 1, Histone H3 N-terminal Peptide, SULFATE ION | | Authors: | Collins, R.E, Horton, J.R, Cheng, X. | | Deposit date: | 2007-11-02 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules.
Nat.Struct.Mol.Biol., 15, 2008
|
|
6A0Q
 
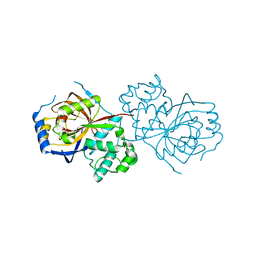 | | The crystal structure of Lpg2622_E64 complex | | Descriptor: | Lpg2622, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE | | Authors: | Gong, X, Ge, H. | | Deposit date: | 2018-06-06 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of the hypothetical protein Lpg2622, a new member of the C1 family peptidases from Legionella pneumophila
FEBS Lett., 592, 2018
|
|
2ZO1
 
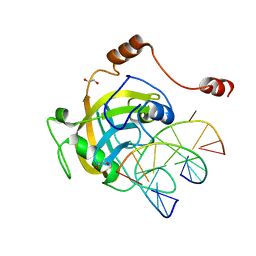 | | Mouse NP95 SRA domain DNA specific complex 2 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*DGP*DTP*DCP*DAP*DGP*(5CM)P*DGP*DCP*DAP*DAP*DTP*DGP*DG)-3'), DNA (5'-D(*DTP*DCP*DCP*DAP*DTP*DGP*DCP*DGP*DCP*DTP*DGP*DAP*DC)-3'), ... | | Authors: | Hashimoto, H, Horton, J.R, Cheng, X. | | Deposit date: | 2008-05-05 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix
Nature, 455, 2008
|
|
7DDN
 
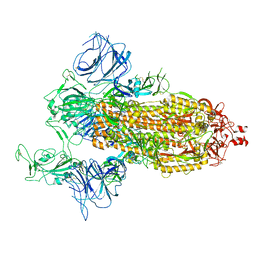 | | SARS-Cov2 S protein at open state | | Descriptor: | Spike glycoprotein | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2020-10-29 | | Release date: | 2020-11-25 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Development and structural basis of a two-MAb cocktail for treating SARS-CoV-2 infections.
Nat Commun, 12, 2021
|
|
7DD2
 
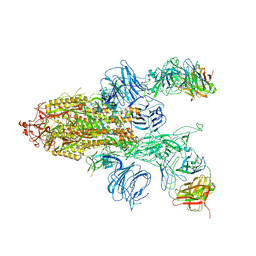 | |
7DK4
 
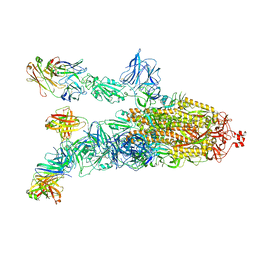 | |
2ZO0
 
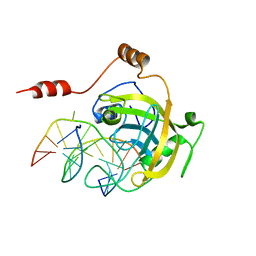 | | mouse NP95 SRA domain DNA specific complex 1 | | Descriptor: | DNA (5'-D(*DGP*DTP*DCP*DAP*DGP*(5CM)P*DGP*DCP*DAP*DAP*DTP*DGP*DG)-3'), DNA (5'-D(*DTP*DCP*DCP*DAP*DTP*DGP*DCP*DGP*DCP*DTP*DGP*DAP*DC)-3'), E3 ubiquitin-protein ligase UHRF1 | | Authors: | Hashimoto, H, Horton, J.R, Cheng, X. | | Deposit date: | 2008-05-05 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix
Nature, 455, 2008
|
|
2ZO2
 
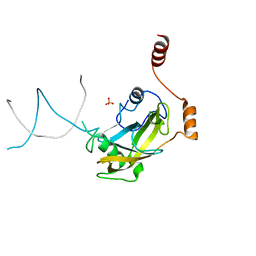 | | Mouse NP95 SRA domain non-specific DNA complex | | Descriptor: | DNA (5'-D(*DAP*DAP*DCP*DTP*DGP*DCP*DGP*DCP*DAP*DGP*DTP*DT)-3'), E3 ubiquitin-protein ligase UHRF1, PHOSPHATE ION | | Authors: | Hashimoto, H, Horton, J.R, Cheng, X. | | Deposit date: | 2008-05-05 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix
Nature, 455, 2008
|
|
6A0N
 
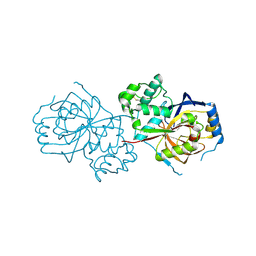 | | The crystal structure of apo-Lpg2622 | | Descriptor: | Lpg2622 | | Authors: | Gong, X, Ge, H. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-12 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the hypothetical protein Lpg2622, a new member of the C1 family peptidases from Legionella pneumophila
FEBS Lett., 592, 2018
|
|
6OOE
 
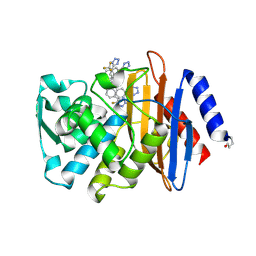 | | CTX-M-27 Beta Lactamase with Compound 20 | | Descriptor: | 3-(1H-tetrazol-5-yl)-N-[3-(1H-tetrazol-5-yl)phenyl]-5-(trifluoromethyl)benzamide, Beta-lactamase | | Authors: | Kemp, M, Chen, Y. | | Deposit date: | 2019-04-23 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | An Empirical Study of Amide-Heteroarene pi-Stacking Interactions Using Reversible Inhibitors of a Bacterial Serine Hydrolase.
Org Chem Front, 6, 2019
|
|
6OOF
 
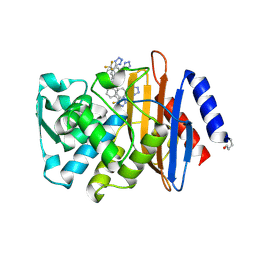 | | CTX-M-14 Beta Lactamase with Compound 20 | | Descriptor: | 3-(1H-tetrazol-5-yl)-N-[3-(1H-tetrazol-5-yl)phenyl]-5-(trifluoromethyl)benzamide, Beta-lactamase | | Authors: | Kemp, M, Chen, Y. | | Deposit date: | 2019-04-23 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.236 Å) | | Cite: | An Empirical Study of Amide-Heteroarene pi-Stacking Interactions Using Reversible Inhibitors of a Bacterial Serine Hydrolase.
Org Chem Front, 6, 2019
|
|
3W0F
 
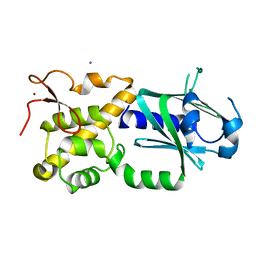 | | Crystal structure of mouse Endonuclease VIII-LIKE 3 (mNEIL3) | | Descriptor: | Endonuclease 8-like 3, IODIDE ION, ZINC ION | | Authors: | Liu, M, Imamura, K, Averill, A.M, Wallace, S.S, Doublie, S. | | Deposit date: | 2012-10-30 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of a Mouse Ortholog of Human NEIL3 with a Marked Preference for Single-Stranded DNA
Structure, 21, 2013
|
|
5HZ6
 
 | |
6OOK
 
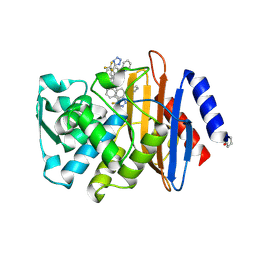 | | CTX-M-14 Beta Lactamase with Compound 3 | | Descriptor: | 3-(pyridin-2-yl)-N-[3-(1H-tetrazol-5-yl)phenyl]-5-(trifluoromethyl)benzamide, Beta-lactamase, PHOSPHATE ION | | Authors: | Kemp, M, Chen, Y. | | Deposit date: | 2019-04-23 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Empirical Study of Amide-Heteroarene pi-Stacking Interactions Using Reversible Inhibitors of a Bacterial Serine Hydrolase.
Org Chem Front, 6, 2019
|
|
5HZ8
 
 | | FABP4_3 in complex with 6,8-Dichloro-4-phenyl-2-piperidin-1-yl-quinoline-3-carboxylic acid | | Descriptor: | 6,8-dichloro-4-phenyl-2-(piperidin-1-yl)quinoline-3-carboxylic acid, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Rudolph, M.G. | | Deposit date: | 2016-02-02 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Design and synthesis of selective, dual fatty acid binding protein 4 and 5 inhibitors.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
5HZ9
 
 | |
6OOJ
 
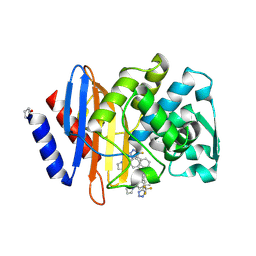 | | CTX-M-14 Beta Lactamase with Compound 14 | | Descriptor: | 3-(1H-pyrazol-1-yl)-N-[3-(1H-tetrazol-5-yl)phenyl]-5-(trifluoromethyl)benzamide, Beta-lactamase | | Authors: | Kemp, M, Chen, Y. | | Deposit date: | 2019-04-23 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Empirical Study of Amide-Heteroarene pi-Stacking Interactions Using Reversible Inhibitors of a Bacterial Serine Hydrolase.
Org Chem Front, 6, 2019
|
|
7RFK
 
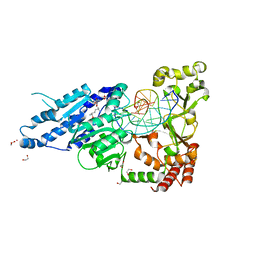 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor Sinefungin | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Repurposing epigenetic inhibitors to target the Clostridioides difficile- specific DNA adenine methyltransferase and sporulation regulator CamA.
Epigenetics, 17, 2022
|
|
7RFM
 
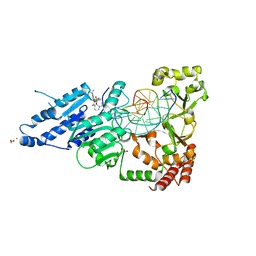 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor EPZ004777 | | Descriptor: | 1,2-ETHANEDIOL, 7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, DNA Strand 1, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Repurposing epigenetic inhibitors to target the Clostridioides difficile- specific DNA adenine methyltransferase and sporulation regulator CamA.
Epigenetics, 17, 2022
|
|
7RFL
 
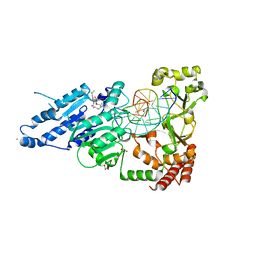 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor SGC0946 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, DNA Strand 1, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Repurposing epigenetic inhibitors to target the Clostridioides difficile- specific DNA adenine methyltransferase and sporulation regulator CamA.
Epigenetics, 17, 2022
|
|
7RFN
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor SGC8158 | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-(4-{[(4'-chloro[1,1'-biphenyl]-3-yl)methyl]amino}butyl)-5'-thioadenosine, DNA Strand 1, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Repurposing epigenetic inhibitors to target the Clostridioides difficile- specific DNA adenine methyltransferase and sporulation regulator CamA.
Epigenetics, 17, 2022
|
|
7BX7
 
 | |
