6L2A
 
 | | A mutant form of M. tb toxin MazEF-mt1 | | Descriptor: | mRNA interferase | | Authors: | Xie, W, Chen, R, Zhou, J. | | Deposit date: | 2019-10-03 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.90044665 Å) | | Cite: | Conserved Conformational Changes in the Regulation ofMycobacterium tuberculosisMazEF-mt1.
Acs Infect Dis., 6, 2020
|
|
6KYS
 
 | | The structure of the M. tb toxin MazF-mt1 | | Descriptor: | Endoribonuclease MazF9 | | Authors: | Xie, W, Chen, R, Zhou, J. | | Deposit date: | 2019-09-20 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.200414 Å) | | Cite: | Conserved Conformational Changes in the Regulation ofMycobacterium tuberculosisMazEF-mt1.
Acs Infect Dis., 6, 2020
|
|
6LM2
 
 | |
6LPS
 
 | |
6M61
 
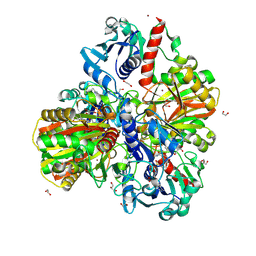 | | Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) with inhibitor heptelidic acid | | Descriptor: | (5aS,6R,9S,9aS)-9-methyl-9-oxidanyl-1-oxidanylidene-6-propan-2-yl-3,5a,6,7,8,9a-hexahydro-2-benzoxepine-4-carboxylic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yan, Y, Zang, X, Cooper, S.J, Lin, H, Zhou, J, Tang, Y. | | Deposit date: | 2020-03-12 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82449543 Å) | | Cite: | Biosynthesis of the fungal glyceraldehyde-3-phosphate dehydrogenase inhibitor heptelidic acid and mechanism of self-resistance
Chem Sci, 11, 2020
|
|
6LKQ
 
 | | The Structural Basis for Inhibition of Ribosomal Translocation by Viomycin | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, L, Wang, Y.H, Lancaster, L, Zhou, J, Noller, H.F. | | Deposit date: | 2019-12-20 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis for inhibition of ribosomal translocation by viomycin.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5WRQ
 
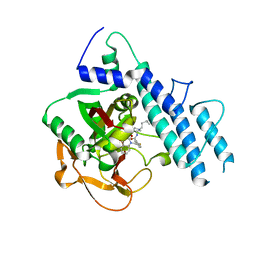 | | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 5-[[2,4-bis(oxidanylidene)quinazolin-1-yl]methyl]-2-fluoranyl-N-[(3R)-1-(3-methylbutyl)pyrrolidin-3-yl]benzamide, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Huang, N, Xu, B.L. | | Deposit date: | 2016-12-03 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor
To Be Published
|
|
5WS0
 
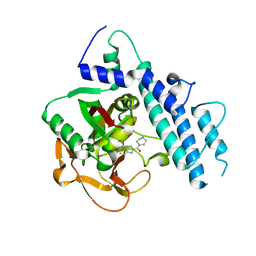 | | Structure of human PARP1 catalytic domain bound to a benzoimidazole inhibitor | | Descriptor: | 2-piperazin-1-ylcarbonyl-1H-benzimidazole-4-carboxamide, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Huang, N, Xu, B.L. | | Deposit date: | 2016-12-04 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a benzoimidazole inhibitor
To Be Published
|
|
5WS1
 
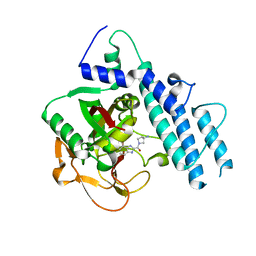 | | Structure of human PARP1 catalytic domain bound to a benzoimidazole inhibitor | | Descriptor: | 2-[(3R)-3-azanylpyrrolidin-1-yl]carbonyl-1H-benzimidazole-4-carboxamide, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Huang, N, Xu, B.L. | | Deposit date: | 2016-12-04 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a benzoimidazole inhibitor
To Be Published
|
|
5WTC
 
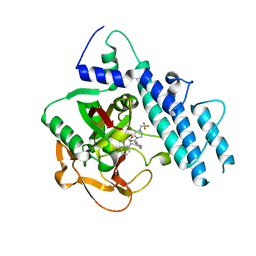 | | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[4-fluoranyl-3-[4-[2,2,2-tris(fluoranyl)ethyl]piperazin-1-yl]carbonyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Huang, N, Xu, B.L. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor
To Be Published
|
|
2RKK
 
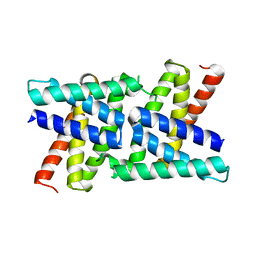 | |
2RKL
 
 | | Crystal Structure of S.cerevisiae Vta1 C-terminal domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Vacuolar protein sorting-associated protein VTA1 | | Authors: | Xiao, J, Xia, H, Zhou, J, Xu, Z. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of vta1 function in the multivesicular body sorting pathway.
Dev.Cell, 14, 2008
|
|
1SPX
 
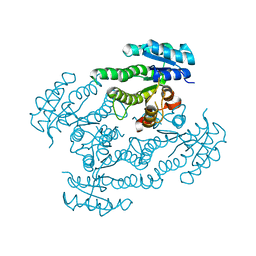 | | Crystal Structure of Glucose Dehydrogenase of Caenorhabditis Elegans in the Apo-Form | | Descriptor: | short-chain reductase family member (5L265) | | Authors: | Schormann, N, Zhou, J, McCombs, D, Bray, T, Symersky, J, Huang, W.-Y, Luan, C.-H, Gray, R, Luo, D, Arabashi, A, Bunzel, B, Nagy, L, Lu, S, Li, S, Lin, G, Zhang, Y, Qiu, S, Tsao, J, Luo, M, Carson, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-17 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Glucose Dehydrogenase of Caenorhabditis Elegans in the Apo-Form: A Member of the SDR-Family
To be Published
|
|
1WNO
 
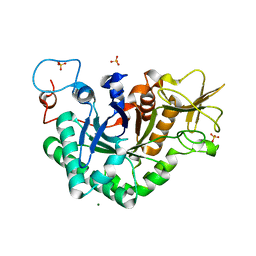 | | Crystal structure of a native chitinase from Aspergillus fumigatus YJ-407 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase, ... | | Authors: | Hu, H, Wang, G, Yang, H, Zhou, J, Mo, L, Yang, K, Jin, C, Jin, C, Rao, Z. | | Deposit date: | 2004-08-07 | | Release date: | 2005-03-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a native chitinase from Aspergillus fumigatus YJ-407
To be Published
|
|
9AUE
 
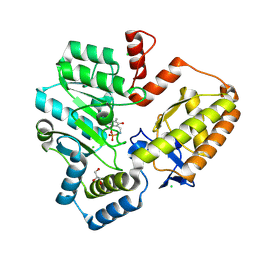 | | Crystal structure of the holo form of GenB2 in complex with PMP | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 6'-epimerase, C-6' aminotransferase, ... | | Authors: | Oliveira, G.S, Bury, P.S, Huang, F, Li, Y, Araujo, N.C, Zhou, J, Sun, Y, Leeper, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2024-02-29 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural and Functional Basis of GenB2 Isomerase Activity from Gentamicin Biosynthesis.
Acs Chem.Biol., 19, 2024
|
|
9AU3
 
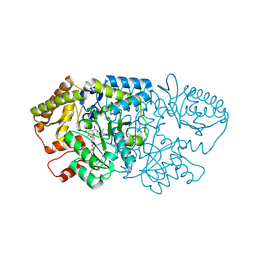 | | Crystal structure of GenB2 in complex with G418 | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 6'-epimerase, C-6' aminotransferase, ... | | Authors: | De Oliveira, G.S, Bury, P.S, Huang, F, Li, Y, Araujo, N.C, Zhou, J, Sun, Y, Leeper, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2024-02-28 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and Functional Basis of GenB2 Isomerase Activity from Gentamicin Biosynthesis.
Acs Chem.Biol., 19, 2024
|
|
7BV5
 
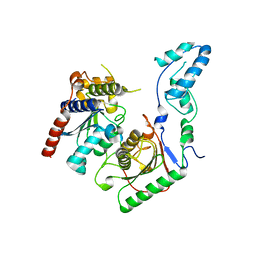 | | Crystal structure of the yeast heterodimeric ADAT2/3 | | Descriptor: | ZINC ION, tRNA-specific adenosine deaminase subunit TAD2, tRNA-specific adenosine deaminase subunit TAD3 | | Authors: | Xie, W, Liu, X, Chen, R, Sun, Y, Chen, R, Zhou, J, Tian, Q. | | Deposit date: | 2020-04-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the yeast heterodimeric ADAT2/3 deaminase.
Bmc Biol., 18, 2020
|
|
1YIS
 
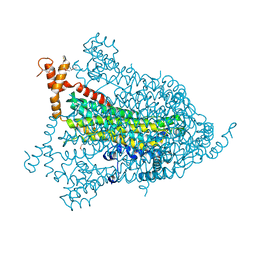 | | Structural genomics of Caenorhabditis elegans: adenylosuccinate lyase | | Descriptor: | SULFATE ION, adenylosuccinate lyase | | Authors: | Symersky, J, Schormann, N, Lu, S, Zhang, Y, Karpova, E, Qiu, S, Huang, W, Cao, Z, Zhou, J, Luo, M, Arabshahi, A, McKinstry, A, Luan, C.-H, Luo, D, Johnson, D, An, J, Tsao, J, Delucas, L, Shang, Q, Gray, R, Li, S, Bray, T, Chen, Y.-J, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-01-12 | | Release date: | 2005-01-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: adenylosuccinate lyase
To be Published
|
|
1XHL
 
 | | Crystal Structure of putative Tropinone Reductase-II from Caenorhabditis Elegans with Cofactor and Substrate | | Descriptor: | 8-METHYL-8-AZABICYCLO[3,2,1]OCTAN-3-ONE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase family member (5L265), ... | | Authors: | Schormann, N, Karpova, E, Zhou, J, Zhang, Y, Symersky, J, Bunzel, R, Huang, W.-Y, Arabshahi, A, Qiu, S, Luan, C.-H, Gray, R, Carson, M, Tsao, J, Luo, M, Johnson, D, Lu, S, Lin, G, Luo, D, Cao, Z, Li, S, McKInstry, A, Shang, Q, Chen, Y.-J, Bray, T, Nagy, L, DeLucas, L, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-20 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of putative Tropinone Reductase-II from Caenorhabditis Elegans with Cofactor and Substrate
To be Published
|
|
1XKQ
 
 | | Crystal Structure of Short-Chain Dehydrogenase/Reductase of unknown Function from Caenorhabditis Elegans with Cofactor | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, short-chain reductase family member (5D234) | | Authors: | Schormann, N, Zhou, J, Karpova, E, Zhang, Y, Symersky, J, Bunzel, B, Huang, W.-Y, Arabshahi, A, Qiu, S, Luan, C.-H, Gray, R, Carson, M, Tsao, J, Luo, M, Johnson, D, Lu, S, Lin, G, Luo, D, Cao, Z, Li, S, McKinstry, A, Shang, Q, Chen, Y.-J, Bray, T, Nagy, L, DeLucas, L, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-29 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Short-Chain Dehydrogenase/Reductase of unknown Function from Caenorhabditis Elegans with Cofactor
To be Published
|
|
3DON
 
 | | Crystal structure of shikimate dehydrogenase from Staphylococcus epidermidis | | Descriptor: | GLYCEROL, Shikimate dehydrogenase | | Authors: | Han, C, Hu, T, Wu, D, Zhou, J, Shen, X, Qu, D, Jiang, H. | | Deposit date: | 2008-07-05 | | Release date: | 2009-05-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystallographic and enzymatic analyses of shikimate dehydrogenase from Staphylococcus epidermidis
Febs J., 276, 2009
|
|
3CM8
 
 | | A RNA polymerase subunit structure from virus | | Descriptor: | Polymerase acidic protein, peptide from RNA-directed RNA polymerase catalytic subunit | | Authors: | He, X, Zhou, J, Zeng, Z, Ma, J, Zhang, R, Rao, Z, Liu, Y. | | Deposit date: | 2008-03-21 | | Release date: | 2008-07-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Crystal structure of the polymerase PAC-PB1N complex from an avian influenza H5N1 virus
Nature, 454, 2008
|
|
4CYE
 
 | | Crystal structure of avian FAK FERM domain FAK31-405 at 3.2A | | Descriptor: | FOCAL ADHESION KINASE 1 | | Authors: | Goni, G.M, Epifano, C, Boskovic, J, Camacho-Artacho, M, Zhou, J, Martin, M.T, Eck, M.J, Kremer, L, Graeter, F, Gervasio, F.L, Perez-Moreno, M, Lietha, D. | | Deposit date: | 2014-04-10 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Phosphatidylinositol 4,5-Bisphosphate Triggers Activation of Focal Adhesion Kinase by Inducing Clustering and Conformational Changes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3DOO
 
 | | Crystal structure of shikimate dehydrogenase from Staphylococcus epidermidis complexed with shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, Shikimate dehydrogenase | | Authors: | Han, C, Hu, T, Wu, D, Zhou, J, Shen, X, Qu, D, Jiang, H. | | Deposit date: | 2008-07-05 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystallographic and enzymatic analyses of shikimate dehydrogenase from Staphylococcus epidermidis
Febs J., 276, 2009
|
|
6LY4
 
 | | The crystal structure of the BM3 mutant LG-23 in complex with testosterone | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional cytochrome P450/NADPH--P450 reductase, IMIDAZOLE, ... | | Authors: | Peng, Y, Chen, J, Zhou, J, Li, A, ReetZ, M.T. | | Deposit date: | 2020-02-13 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Regio- and Stereoselective Steroid Hydroxylation at C7 by Cytochrome P450 Monooxygenase Mutants.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
