5BU3
 
 | | Crystal Structure of Diels-Alderase PyrI4 in complex with its product | | Descriptor: | (4S,4aS,6aS,8R,9R,10aR,13R,14aS,18aR,18bR)-9-ethyl-4,8,19-trihydroxy-10a,12,13,18a-tetramethyl-2,3,4,4a,5,6,6a,7,8,9,10,10a,13,14,18a,18b-hexadecahydro-1H-14a,17-(metheno)benzo[b]naphtho[2,1-h]azacyclododecine-16,18(15H,17H)-dione, GLYCEROL, PyrI4 | | Authors: | Pan, L, Guo, Y, Liu, J. | | Deposit date: | 2015-06-03 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Enzyme-Dependent [4 + 2] Cycloaddition Depends on Lid-like Interaction of the N-Terminal Sequence with the Catalytic Core in PyrI4
Cell Chem Biol, 23, 2016
|
|
2IEQ
 
 | |
5BTU
 
 | |
6AJ2
 
 | | The structure of ICAM-5 triggered Enterovirus D68 virus A-particle | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
6AJ0
 
 | | The structure of Enterovirus D68 mature virion | | Descriptor: | Capsid protein VP3, Capsid protein VP4, Viral protein 1, ... | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-25 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
6AJ9
 
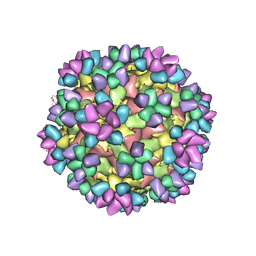 | | The structure of Enterovirus D68 mature virion in complex with Fab 15C5 and 11G1 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-27 | | Release date: | 2018-11-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
6AJ3
 
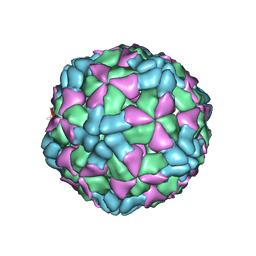 | | The structure of Enterovirus D68 procapsid | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
6AJ7
 
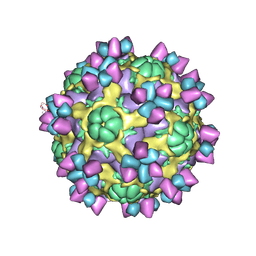 | | The structure of Enterovirus D68 mature virion in complex with Fab 15C5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-27 | | Release date: | 2018-11-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
5XGV
 
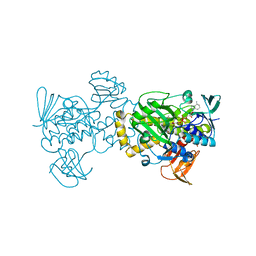 | |
5ZFM
 
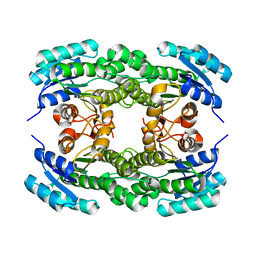 | | Ketoreductase LbCR mutant - M6 | | Descriptor: | 3-oxoacyl-acyl carrier protein reductase | | Authors: | Gong, X.M, Zheng, G.W, Xu, J.H. | | Deposit date: | 2018-03-06 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of an Engineered Ketoreductase with Simultaneously Improved Thermostability and Activity for Making a Bulky Atorvastatin Precursor
Acs Catalysis, 9, 2019
|
|
5ZI0
 
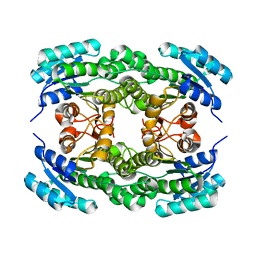 | | Ketoreductase LbCR mutant - M8 | | Descriptor: | 3-oxoacyl-acyl carrier protein reductase | | Authors: | Gong, X.M, Zheng, G.W, Xu, J.H. | | Deposit date: | 2018-03-14 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Development of an Engineered Ketoreductase with Simultaneously Improved Thermostability and Activity for Making a Bulky Atorvastatin Precursor
Acs Catalysis, 9, 2019
|
|
7Y1T
 
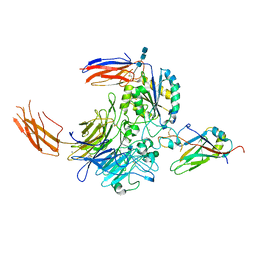 | | Complex of integrin alphaV/beta8 and L-TGF-beta1 at a ratio of 1:2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duan, Z, Zhang, Z. | | Deposit date: | 2022-06-08 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Specificity of TGF-beta 1 signal designated by LRRC33 and integrin alpha V beta 8.
Nat Commun, 13, 2022
|
|
7Y1R
 
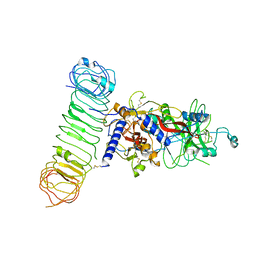 | | Human L-TGF-beta1 in complex with the anchor protein LRRC33 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duan, Z, Zhang, Z. | | Deposit date: | 2022-06-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Specificity of TGF-beta 1 signal designated by LRRC33 and integrin alpha V beta 8.
Nat Commun, 13, 2022
|
|
5JB1
 
 | | Pseudo-atomic structure of Human Papillomavirus Type 59 L1 Virus-like Particle | | Descriptor: | Major capsid protein L1 | | Authors: | Li, Z.H, Yan, X.D, Yu, H, Zheng, Q.B, Gu, Y, Li, S.W. | | Deposit date: | 2016-04-13 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | The C-Terminal Arm of the Human Papillomavirus Major Capsid Protein Is Immunogenic and Involved in Virus-Host Interaction.
Structure, 24, 2016
|
|
5XS5
 
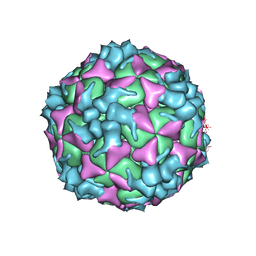 | | Structure of Coxsackievirus A6 (CVA6) virus procapsid particle | | Descriptor: | Genome polyprotein | | Authors: | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Cheng, T, Li, S.W. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
5XS7
 
 | | Structure of Coxsackievirus A6 (CVA6) virus A-particle in complex with the neutralizing antibody fragment 1D5 | | Descriptor: | Genome polyprotein, Heavy chain of Fab 1D5, Light chain of Fab 1D5 | | Authors: | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Li, S.W, Cheng, T. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
5J6R
 
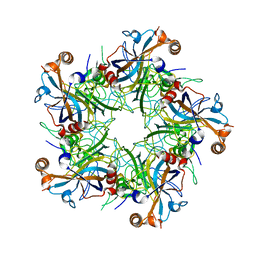 | | Crystal structure of Human Papillomavirus Type 59 L1 pentamer | | Descriptor: | Major capsid protein L1 | | Authors: | Li, Z.H, Yan, X.D, Yu, H, Gu, Y, Li, S.W. | | Deposit date: | 2016-04-05 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.011 Å) | | Cite: | The C-Terminal Arm of the Human Papillomavirus Major Capsid Protein Is Immunogenic and Involved in Virus-Host Interaction.
Structure, 24, 2016
|
|
8JTL
 
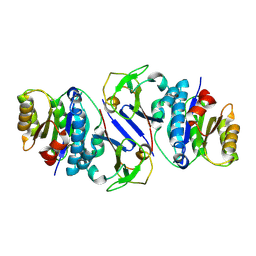 | | Structure of OY phytoplasma SAP05 binding with AtRpn10 | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4 homolog, Sequence-variable mosaic (SVM) signal sequence domain-containing protein | | Authors: | Du, Y.X, Zhang, L.Y, Zheng, Q.Y. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure basis for recognition of plant Rpn10 by phytoplasma SAP05 in ubiquitin-independent protein degradation.
Iscience, 27, 2024
|
|
8JTK
 
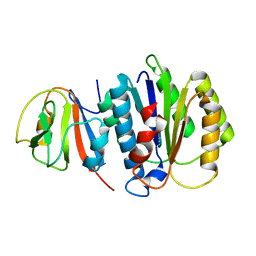 | | Structure of AYWB phytoplasma SAP05 recognizing AtRpn10 | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4 homolog, Sequence-variable mosaic (SVM) signal sequence domain-containing protein | | Authors: | Du, Y.X, Zhang, L.Y, Zheng, Q.Y. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure basis for recognition of plant Rpn10 by phytoplasma SAP05 in ubiquitin-independent protein degradation.
Iscience, 27, 2024
|
|
2R2V
 
 | |
3CRP
 
 | | A heterospecific leucine zipper tetramer | | Descriptor: | GCN4 leucine zipper, SODIUM ION | | Authors: | Liu, J. | | Deposit date: | 2008-04-07 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A heterospecific leucine zipper tetramer.
Chem.Biol., 15, 2008
|
|
6LHB
 
 | | The cryo-EM structure of coxsackievirus A16 A-particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHC
 
 | | The cryo-EM structure of coxsackievirus A16 empty particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHL
 
 | | The cryo-EM structure of coxsackievirus A16 A-particle in complex with Fab 18A7 | | Descriptor: | VP1 protein, VP2 protein, VP3 protein | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHP
 
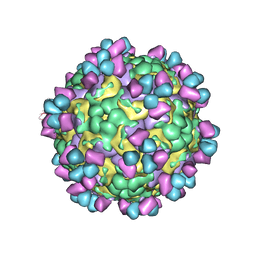 | | The cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab 14B10 | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
