6KJP
 
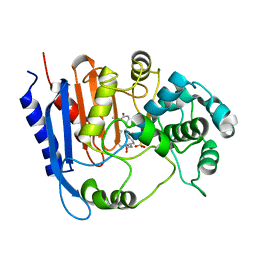 | | Functional and structural insights into the unusual oxyanion hole-like geometry in macrolactin acyltransferase selective for dicarboxylic acyl donors | | Descriptor: | (3~{Z},5~{E},8~{S},9~{E},11~{E},14~{S},16~{R},17~{Z},19~{E},24~{R})-24-methyl-8,14,16-tris(oxidanyl)-1-oxacyclotetracosa-3,5,9,11,17,19-hexaen-2-one, Putative beta-lactamase, SULFATE ION | | Authors: | Xiao, F, Dong, S, Feng, Y, Li, W. | | Deposit date: | 2019-07-23 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural Basis of Specificity for Carboxyl-Terminated Acyl Donors in a Bacterial Acyltransferase.
J.Am.Chem.Soc., 142, 2020
|
|
6KJJ
 
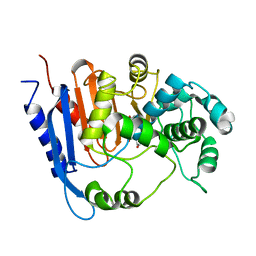 | | Functional and structural insights into the unusual oxyanion hole-like geometry in macrolactin acyltransferase selective for dicarboxylic acyl donors | | Descriptor: | 4-(2-acetamidoethylsulfanyl)-4-oxidanylidene-butanoic acid, Putative beta-lactamase | | Authors: | Xiao, F, Dong, S, Feng, Y, Li, W. | | Deposit date: | 2019-07-22 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | Structural Basis of Specificity for Carboxyl-Terminated Acyl Donors in a Bacterial Acyltransferase.
J.Am.Chem.Soc., 142, 2020
|
|
6KJH
 
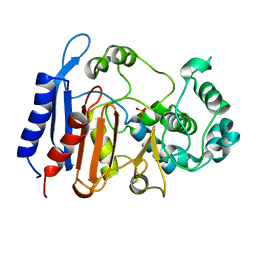 | | Functional and structural insights into the unusual oxyanion hole-like geometry in macrolactin acyltransferase selective for dicarboxylic acyl donors | | Descriptor: | Putative beta-lactamase, SULFATE ION | | Authors: | Xiao, F, Sheng, D, Feng, Y, Li, W. | | Deposit date: | 2019-07-22 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Basis of Specificity for Carboxyl-Terminated Acyl Donors in a Bacterial Acyltransferase.
J.Am.Chem.Soc., 142, 2020
|
|
6KJQ
 
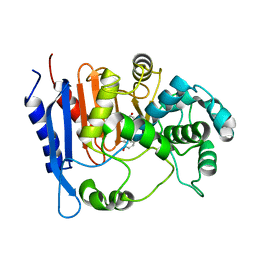 | | Functional and structural insights into the unusual oxyanion hole-like geometry in macrolactin acyltransferase selective for dicarboxylic acyl donors | | Descriptor: | (3~{Z},5~{E},8~{S},9~{E},11~{E},14~{S},16~{R},17~{Z},19~{E},24~{R})-24-methyl-8,14,16-tris(oxidanyl)-1-oxacyclotetracosa-3,5,9,11,17,19-hexaen-2-one, Putative beta-lactamase | | Authors: | Xiao, F, Dong, S, Feng, Y, Li, W. | | Deposit date: | 2019-07-23 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Specificity for Carboxyl-Terminated Acyl Donors in a Bacterial Acyltransferase.
J.Am.Chem.Soc., 142, 2020
|
|
6L5R
 
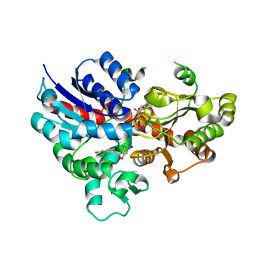 | | crystal structure of GgCGT in complex with UDP-Glu | | Descriptor: | 3-(4-HYDROXYPHENYL)-1-(2,4,6-TRIHYDROXYPHENYL)PROPAN-1-ONE, GgCGT, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhang, M, Li, F.D, Ye, M. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Functional Characterization and Structural Basis of an Efficient Di-C-glycosyltransferase fromGlycyrrhiza glabra.
J.Am.Chem.Soc., 142, 2020
|
|
4R6V
 
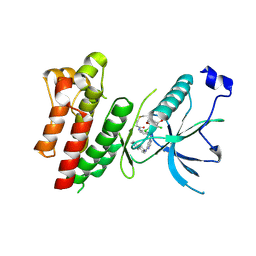 | | Crystal Structure of FGF Receptor (FGFR) 4 Kinase Harboring the V550L Gate-Keeper Mutation in Complex with FIIN-3, an Irreversible Tyrosine Kinase Inhibitor Capable of Overcoming FGFR kinase Gate-Keeper Mutations | | Descriptor: | Fibroblast growth factor receptor 4, N-[4-({[(2,6-dichloro-3,5-dimethoxyphenyl)carbamoyl](6-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino}methyl)phenyl]propanamide, SULFATE ION | | Authors: | Huang, Z, Mohammadi, M. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5GMP
 
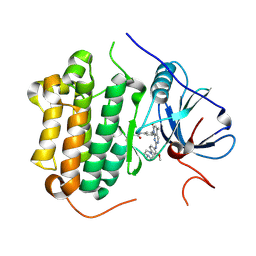 | | Crystal structure of EGFR 696-1022 T790M in complex with XTF-262 | | Descriptor: | Epidermal growth factor receptor, N-[3-[2-[[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino]-5-methyl-7-oxidanylidene-pyrido[2,3-d]pyrimidin-8-yl]phenyl]prop-2-enamide | | Authors: | Yan, X.E, Yun, C.H. | | Deposit date: | 2016-07-14 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | A structure-guided optimization of pyrido[2,3-d]pyrimidin-7-ones as selective inhibitors of EGFR(L858R/T790M) mutant with improved pharmacokinetic properties.
Eur J Med Chem, 126, 2017
|
|
4FXB
 
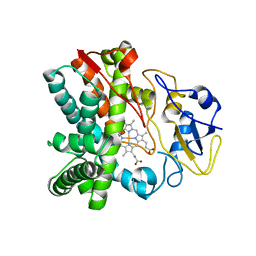 | | Crystal structure of CYP105N1 from Streptomyces coelicolor: a cytochrome P450 oxidase in the coelibactin siderophore biosynthetic pathway | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 | | Authors: | Hong, M.K, Lim, Y.R, Kim, J.K, Kim, D.H, Kang, L.W. | | Deposit date: | 2012-07-03 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of cytochrome P450 CYP105N1 from Streptomyces coelicolor, an oxidase in the coelibactin siderophore biosynthetic pathway
Arch.Biochem.Biophys., 528, 2012
|
|
4IM2
 
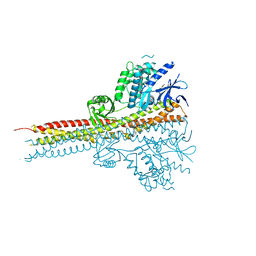 | | Structure of Tank-Binding Kinase 1 | | Descriptor: | CHLORIDE ION, N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, Serine/threonine-protein kinase TBK1 | | Authors: | Tu, D, Eck, M.J. | | Deposit date: | 2013-01-01 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5001 Å) | | Cite: | Structure and ubiquitination-dependent activation of TANK-binding kinase 1.
Cell Rep, 3, 2013
|
|
4IM0
 
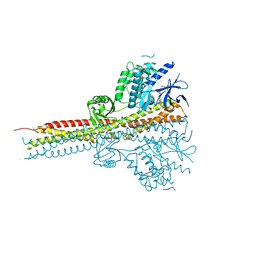 | | Structure of Tank-Binding Kinase 1 | | Descriptor: | N-{3-[(5-cyclopropyl-2-{[3-(morpholin-4-ylmethyl)phenyl]amino}pyrimidin-4-yl)amino]propyl}cyclobutanecarboxamide, Serine/threonine-protein kinase TBK1 | | Authors: | Tu, D, Eck, M.J. | | Deposit date: | 2013-01-01 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4001 Å) | | Cite: | Structure and ubiquitination-dependent activation of TANK-binding kinase 1.
Cell Rep, 3, 2013
|
|
4IM3
 
 | | Structure of Tank-Binding Kinase 1 | | Descriptor: | CHLORIDE ION, MERCURY (II) ION, N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, ... | | Authors: | Tu, D, Eck, M.J. | | Deposit date: | 2013-01-01 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.342 Å) | | Cite: | Structure and ubiquitination-dependent activation of TANK-binding kinase 1.
Cell Rep, 3, 2013
|
|
4LGH
 
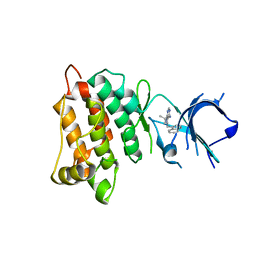 | |
4LGG
 
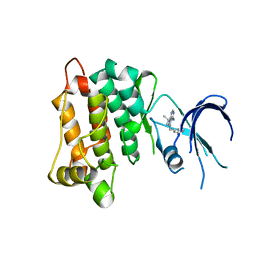 | | Structure of 3MB-PP1 bound to analog-sensitive Src kinase | | Descriptor: | 1-tert-butyl-3-(3-methylbenzyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Lopez, M.S, Dar, A.C, Shokat, K.M. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure-guided inhibitor design expands the scope of analog-sensitive kinase technology.
Acs Chem.Biol., 8, 2013
|
|
6JOL
 
 | |
6JOI
 
 | |
6JOK
 
 | | Crystal structure of PDGFRA in complex with sunitinib by soaking | | Descriptor: | CHLORIDE ION, N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide, Platelet-derived growth factor receptor alpha | | Authors: | Liang, L, Yan, X.E, Yun, C.H. | | Deposit date: | 2019-03-22 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of PDGFRA in complex with sunitinib by soaking
To Be Published
|
|
6JOJ
 
 | |
2HWO
 
 | |
2HWP
 
 | |
4QQC
 
 | | Crystal Structure of FGF Receptor (FGFR) 4 Kinase Domain in Complex with FIIN-2, an Irreversible Tyrosine Kinase Inhibitor Capable of Overcoming FGFR Kinase Gate-Keeper Mutations | | Descriptor: | Fibroblast growth factor receptor 4, N-(4-{[3-(3,5-dimethoxyphenyl)-7-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-oxo-3,4-dihydropyrimido[4,5-d]pyrimidin-1(2H)-yl]methyl}phenyl)propanamide, SULFATE ION | | Authors: | Huang, Z, Mohammadi, M. | | Deposit date: | 2014-06-27 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
