6FLR
 
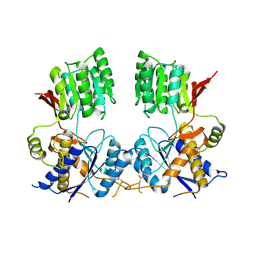 | | Super-open structure of the AMPAR GluA3 N-terminal domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 3 | | Authors: | Garcia-Nafria, J. | | Deposit date: | 2018-01-27 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Druggability Simulations and X-Ray Crystallography Reveal a Ligand-Binding Site in the GluA3 AMPA Receptor N-Terminal Domain.
Structure, 27, 2019
|
|
6FPJ
 
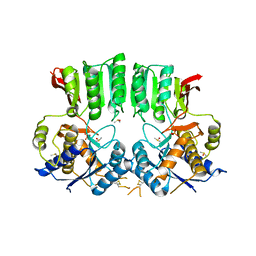 | | Structure of the AMPAR GluA3 N-terminal domain bound to phosphate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Herguedas, B, Garcia-Nafria, J, Greger, I. | | Deposit date: | 2018-02-09 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Druggability Simulations and X-Ray Crystallography Reveal a Ligand-Binding Site in the GluA3 AMPA Receptor N-Terminal Domain.
Structure, 27, 2019
|
|
8K05
 
 | |
8K07
 
 | |
8K06
 
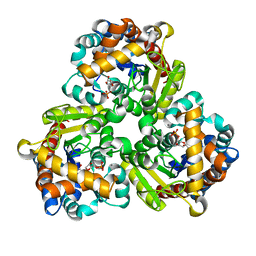 | | Pseudouridine 5'-monophosphate glycosylase from Arabidopsis thaliana -- PSU, R5P bound K185A mutant | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, MANGANESE (II) ION, PSEUDOURIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Lee, J.Y, Kim, S.H, Rhee, S.K. | | Deposit date: | 2023-07-07 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Structure and function of the pseudouridine 5'-monophosphate glycosylase PUMY from Arabidopsis thaliana.
Rna Biol., 21, 2024
|
|
5DO7
 
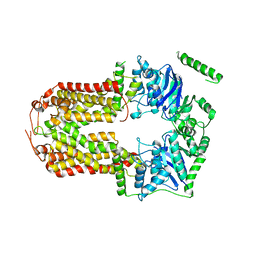 | | Crystal Structure of the Human Sterol Transporter ABCG5/ABCG8 | | Descriptor: | ATP-binding cassette sub-family G member 5, ATP-binding cassette sub-family G member 8 | | Authors: | Lee, J.-Y, Kinch, L.N, Borek, D.M, Urbatsch, I.L, Xie, X.-S, Grishin, N.V, Cohen, J.C, Otwinowski, Z, Hobbs, H.H, Rosenbaum, D.M. | | Deposit date: | 2015-09-10 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.93 Å) | | Cite: | Crystal structure of the human sterol transporter ABCG5/ABCG8.
Nature, 533, 2016
|
|
6JC2
 
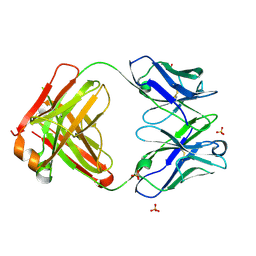 | | Crystal structure of the Fab fragment of ipilimumab | | Descriptor: | SULFATE ION, ipilimumab fab heavy chain, ipilimumab fab light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2019-01-27 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of the Fab Fragment of an Anti-CTLA-4 Antibody, Ipilimumab, Used for Cancer Immunotherapy
Bull.Korean Chem.Soc., 40, 2019
|
|
4MGF
 
 | |
4MF9
 
 | |
3KK6
 
 | | Crystal Structure of Cyclooxygenase-1 in complex with celecoxib | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[5-(4-METHYLPHENYL)-3-(TRIFLUOROMETHYL)-1H-PYRAZOL-1-YL]BENZENESULFONAMIDE, CITRATE ANION, ... | | Authors: | Sidhu, R.S. | | Deposit date: | 2009-11-04 | | Release date: | 2009-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Coxibs interfere with the action of aspirin by binding tightly to one monomer of cyclooxygenase-1.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2B5S
 
 | | Crystal structure of peach Pru p3, the prototypic member of the family of plant non-specific lipid transfer protein pan-allergens | | Descriptor: | HEPTANE, LAURIC ACID, Non-specific lipid transfer protein, ... | | Authors: | Pasquato, N, Berni, R, Folli, C, Folloni, S, Cianci, M, Pantano, S, Helliwell, R.J, Zanotti, G. | | Deposit date: | 2005-09-29 | | Release date: | 2005-11-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Peach Pru p 3, the Prototypic Member of the Family of Plant Non-specific Lipid Transfer Protein Pan-allergens
J.Mol.Biol., 356, 2006
|
|
6JKG
 
 | | The NAD+-free form of human NSDHL | | Descriptor: | Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating | | Authors: | Kim, D, Lee, S.J, Lee, B. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of human NSDHL and development of its novel inhibitor with the potential to suppress EGFR activity.
Cell.Mol.Life Sci., 78, 2021
|
|
6JKH
 
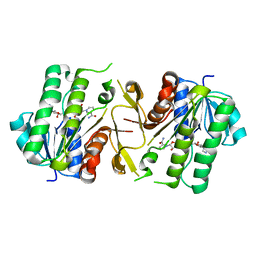 | | The NAD+-bound form of human NSDHL | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating | | Authors: | Kim, D, Lee, S.J, Lee, B. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of human NSDHL and development of its novel inhibitor with the potential to suppress EGFR activity.
Cell.Mol.Life Sci., 78, 2021
|
|
3UY5
 
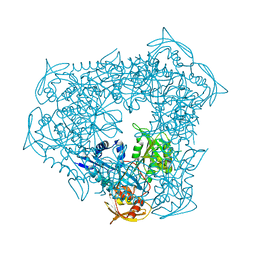 | | crystal structure of Eis from Mycobacterium tuberculosis | | Descriptor: | Enhanced intracellular survival protein | | Authors: | Kim, K.H, Suh, S.W. | | Deposit date: | 2011-12-05 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7
Proc.Natl.Acad.Sci.USA, 2012
|
|
1W01
 
 | |
7MSO
 
 | |
8GTG
 
 | | Corticotropin-releasing hormone receptor 1(CRF1R) bound with BMK-I-152 by XFEL | | Descriptor: | 8-(4-bromanyl-2,6-dimethoxy-phenyl)-~{N},~{N}-bis(2-methoxyethyl)-2,7-dimethyl-pyrazolo[1,5-a][1,3,5]triazin-4-amine, Endolysin, Isoform CRF-R2 of Corticotropin-releasing factor receptor 1 | | Authors: | Cho, H.S, Kim, H. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser.
Exp.Mol.Med., 55, 2023
|
|
8GTI
 
 | | Corticotropin-releasing hormone receptor 1(CRF1R) bound with BMK-C205 by XFEL | | Descriptor: | 8-(4-bromanyl-2,6-dimethoxy-phenyl)-~{N}-butyl-~{N}-(cyclopropylmethyl)-2,7-dimethyl-pyrazolo[1,5-a][1,3,5]triazin-4-amine, Endolysin, Isoform CRF-R2 of Corticotropin-releasing factor receptor 1, ... | | Authors: | Cho, H.S, Kim, H. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser.
Exp.Mol.Med., 55, 2023
|
|
8GTM
 
 | | Corticotropin-releasing hormone receptor 1(CRF1R) bound with BMK-C203 by XFEL | | Descriptor: | 7-(4-bromanyl-2,6-dimethoxy-phenyl)-4,8-dimethyl-~{N},~{N}-bis[4,4,4-tris(fluoranyl)butyl]-1$l^{4},3,5,9-tetrazabicyclo[4.3.0]nona-1(6),2,4,8-tetraen-2-amine, Endolysin, Isoform CRF-R2 of Corticotropin-releasing factor receptor 1 | | Authors: | Cho, H.S, Kim, H. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser.
Exp.Mol.Med., 55, 2023
|
|
1VZZ
 
 | |
1W02
 
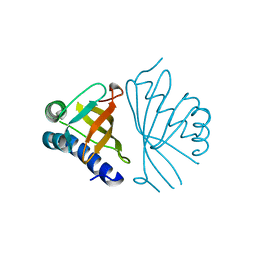 | |
1W00
 
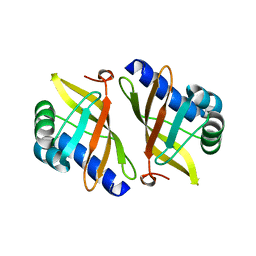 | | Crystal structure of mutant enzyme D103L of Ketosteroid Isomerase from Pseudomonas putida biotype B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 2004-05-30 | | Release date: | 2005-05-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Double-Mutant Cycle Analysis of a Hydrogen Bond Network in Ketosteroid Isomerase from Pseudomonas Putida Biotype B
Biochem.J., 382, 2004
|
|
2KI2
 
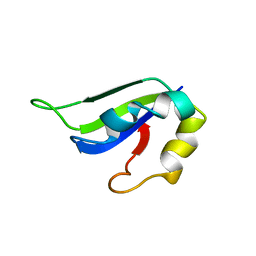 | | Solution Structure of ss-DNA Binding Protein 12RNP2 Precursor, HP0827(O25501_HELPY) form Helicobacter pylori | | Descriptor: | Ss-DNA binding protein 12RNP2 | | Authors: | Ma, C, Lee, J, Kim, J, Park, S, Kwon, A, Lee, B. | | Deposit date: | 2009-04-20 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of HP0827 (O25501_HELPY) from Helicobacter pylori: model of the possible RNA-binding site
J.Biochem., 146, 2009
|
|
7XCL
 
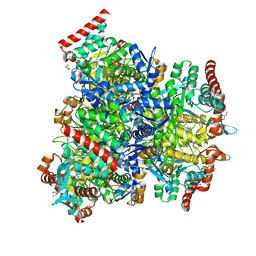 | |
7XCM
 
 | | Crystal structure of sulfite MttB structure at 3.2 A resolution | | Descriptor: | 3-METHYL-5-SULFO-PYRROLIDINE-2-CARBOXYLIC ACID, GLYCEROL, SODIUM ION, ... | | Authors: | Li, J, Chan, M.K. | | Deposit date: | 2022-03-24 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Insights into pyrrolysine function from structures of a trimethylamine methyltransferase and its corrinoid protein complex.
Commun Biol, 6, 2023
|
|
