6JCE
 
 | | NMR solution and X-ray crystal structures of a DNA containing both right-and left-handed parallel-stranded G-quadruplexes | | Descriptor: | 29-mer DNA | | Authors: | Winnerdy, F.R, Bakalar, B, Maity, A, Vandana, J.J, Mechulam, Y, Schmitt, E, Phan, A.T. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution and X-ray crystal structures of a DNA molecule containing both right- and left-handed parallel-stranded G-quadruplexes.
Nucleic Acids Res., 47, 2019
|
|
4Q24
 
 | | Crystal structure of Cyclo(L-leucyl-L-phenylalanyl) synthase | | Descriptor: | Cyclo(L-leucyl-L-phenylalanyl) synthase, PHENYLMETHYL N-[(2S)-4-CHLORO-3-OXO-1-PHENYL-BUTAN-2-YL]CARBAMATE | | Authors: | Moutiez, M, Schmitt, E, Seguin, J, Thai, R, Favry, E, Mechulam, Y, Gondry, M. | | Deposit date: | 2014-04-07 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Unravelling the mechanism of non-ribosomal peptide synthesis by cyclodipeptide synthases.
Nat Commun, 5, 2014
|
|
4MO0
 
 | |
8PHV
 
 | | Structure of Human selenomethionylated Cdc123 bound to domain 3 of eIF2 gamma | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division cycle protein 123 homolog, Eukaryotic translation initiation factor 2 subunit 3 | | Authors: | Schmitt, E, Mechulam, Y, Cardenal Peralta, C, Fagart, J, Seufert, W. | | Deposit date: | 2023-06-20 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Binding of human Cdc123 to eIF2 gamma.
J.Struct.Biol., 215, 2023
|
|
8PHD
 
 | | Structure of Human Cdc123 bound to domain 3 of eIF2 gamma and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division cycle protein 123 homolog, Eukaryotic translation initiation factor 2 subunit 3, ... | | Authors: | Schmitt, E, Mechulam, Y, Cardenal Peralta, C, Fagart, J, Seufert, W. | | Deposit date: | 2023-06-19 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Binding of human Cdc123 to eIF2 gamma.
J.Struct.Biol., 215, 2023
|
|
3PMV
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | GLUTAMIC ACID, GLYCEROL, Glutamate receptor 2, ... | | Authors: | Maclean, J.K.F, Jamieson, C, Brown, C.I, Campbell, R.A, Gillen, K.J, Gillespie, J, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure based evolution of a novel series of positive modulators of the AMPA receptor.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3PMX
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, GLUTAMIC ACID, ... | | Authors: | Maclean, J.K.F, Jamieson, C, Brown, C.I, Campbell, R.A, Gillen, K.J, Gillespie, J, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-12 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure based evolution of a novel series of positive modulators of the AMPA receptor.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3O6I
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | 2-[({3-tert-butyl-4-[(methylamino)methyl]-1H-pyrazol-1-yl}acetyl)amino]-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxamide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Maclean, J.K.F, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Martin, F, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-29 | | Release date: | 2010-09-15 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: structure-based lead optimization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3O29
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | GLUTAMIC ACID, GLYCEROL, Glutamate receptor 2, ... | | Authors: | Maclean, J.K.F, Basten, S, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Moir, E.M, Morrow, J.A, Papakosta, M, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: discovery and structure based hit-to-lead studies.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3O28
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | 2-({[3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl]acetyl}amino)-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxamide, DIMETHYL SULFOXIDE, GLUTAMIC ACID, ... | | Authors: | Maclean, J.K.F, Basten, S, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Moir, E.M, Morrow, J.A, Papakosta, M, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: discovery and structure based hit-to-lead studies.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7YZN
 
 | | Structure of C-terminally truncated aIF5B from Pyrococcus abyssi complexed with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Probable translation initiation factor IF-2, ... | | Authors: | Bourgeois, G, Schmitt, E, Mechulam, Y, Coureux, P.D, Kazan, R. | | Deposit date: | 2022-02-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Role of aIF5B in archaeal translation initiation.
Nucleic Acids Res., 50, 2022
|
|
7YYP
 
 | | Structure of aIF5B from Pyrococcus abyssi complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, NITRATE ION, Probable translation initiation factor IF-2 | | Authors: | Bourgeois, G, Schmitt, E, Mechulam, Y, Coureux, P.D, Kazan, R. | | Deposit date: | 2022-02-18 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Role of aIF5B in archaeal translation initiation.
Nucleic Acids Res., 50, 2022
|
|
7ZAI
 
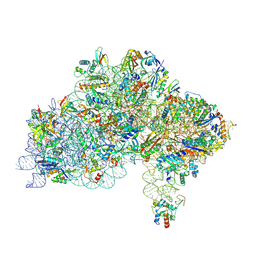 | | Cryo-EM structure of a Pyrococcus abyssi 30S bound to Met-initiator tRNA, mRNA and aIF1A. | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Coureux, P.D, Bourgeois, G, Mechulam, Y, Schmitt, E, Kazan, R. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-13 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Role of aIF5B in archaeal translation initiation.
Nucleic Acids Res., 50, 2022
|
|
3O6H
 
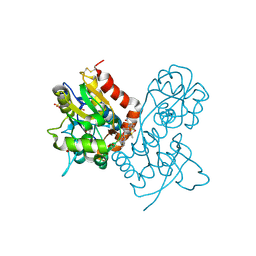 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | 1,2-ETHANEDIOL, 2-[({4-[(ethylamino)methyl]-3-(trifluoromethyl)-1H-pyrazol-1-yl}acetyl)amino]-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Maclean, J.K.F, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Martin, F, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-29 | | Release date: | 2010-09-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: structure-based lead optimization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7ZAG
 
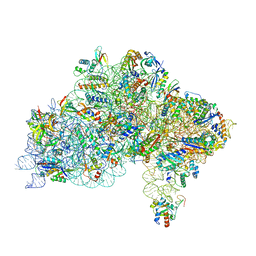 | | Cryo-EM structure of a Pyrococcus abyssi 30S bound to Met-initiator tRNA,mRNA, aIF1A and the C-terminal domain of aIF5B. | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Coureux, P.D, Bourgeois, G, Mechulam, Y, Schmitt, E, Kazan, R. | | Deposit date: | 2022-03-22 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Role of aIF5B in archaeal translation initiation.
Nucleic Acids Res., 50, 2022
|
|
7ZAH
 
 | | Cryo-EM structure of a Pyrococcus abyssi 30S bound to Met-initiator tRNA, mRNA, aIF1A and aIF5B | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Coureux, P.D, Bourgeois, G, Mechulam, Y, Schmitt, E, Kazan, R. | | Deposit date: | 2022-03-22 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Role of aIF5B in archaeal translation initiation.
Nucleic Acids Res., 50, 2022
|
|
3O6G
 
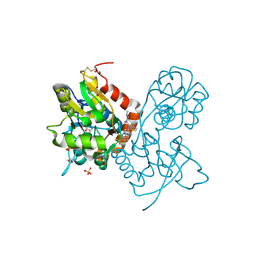 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Maclean, J.K.F, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Martin, F, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-29 | | Release date: | 2010-09-15 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: structure-based lead optimization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3PMW
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | DIMETHYL SULFOXIDE, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Maclean, J.K.F, Jamieson, C, Brown, C.I, Campbell, R.A, Gillen, K.J, Gillespie, J, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-12 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure based evolution of a novel series of positive modulators of the AMPA receptor.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4ZBM
 
 | | Crystal structure of Drosophila cyclic nucleotide gated channel pore mimicking NaK mutant | | Descriptor: | BARIUM ION, POTASSIUM ION, Potassium channel protein | | Authors: | Lam, Y, Zeng, W, Derebe, M.G, Jiang, Y. | | Deposit date: | 2015-04-14 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural implications of weak Ca2+ block in Drosophila cyclic nucleotide-gated channels.
J.Gen.Physiol., 146, 2015
|
|
3KMG
 
 | |
2VH7
 
 | |
2D6F
 
 | |
1EG0
 
 | | FITTING OF COMPONENTS WITH KNOWN STRUCTURE INTO AN 11.5 A CRYO-EM MAP OF THE E.COLI 70S RIBOSOME | | Descriptor: | FORMYL-METHIONYL-TRNA, FRAGMENT OF 16S RRNA HELIX 23, FRAGMENT OF 23S RRNA, ... | | Authors: | Gabashvili, I.S, Agrawal, R.K, Spahn, C.M.T, Grassucci, R.A, Svergun, D.I, Frank, J, Penczek, P. | | Deposit date: | 2000-02-11 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Solution structure of the E. coli 70S ribosome at 11.5 A resolution.
Cell(Cambridge,Mass.), 100, 2000
|
|
6DML
 
 | | A multiconformer ligand model of 3,5 dimethylisoxaxole bound to the bromodomain of human BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 4-((2-(tert-butyl)phenyl)amino)-7-(3,5-dimethylisoxazol-4-yl)-6-methoxy-1,5-naphthyridine-3-carboxylic acid, Bromodomain-containing protein 4 | | Authors: | Hudson, B.M, van Zundert, G, Keedy, D.A, Fonseca, R, Heliou, A, Suresh, P, Borrelli, K, Day, T, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.
J. Med. Chem., 61, 2018
|
|
1MEA
 
 | |
