6J10
 
 | | Ciclopirox inhibits Hepatitis B Virus secretion by blocking capsid assembly | | Descriptor: | 6-cyclohexyl-4-methyl-1-oxidanyl-pyridin-2-one, Capsid protein | | Authors: | Park, S, Jin, M.S, Cho, Y, Kang, J, Kim, S, Park, M, Park, H, Kim, J, Park, S, Hwang, J, Kim, Y, Kim, Y.J. | | Deposit date: | 2018-12-27 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ciclopirox inhibits Hepatitis B Virus secretion by blocking capsid assembly.
Nat Commun, 10, 2019
|
|
2NWG
 
 | | Structure of CXCL12:heparin disaccharide complex | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Stromal cell-derived factor 1 | | Authors: | Murphy, J.W, Cho, Y, Lolis, E. | | Deposit date: | 2006-11-14 | | Release date: | 2007-02-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural and Functional Basis of CXCL12 (Stromal Cell-derived Factor-1{alpha}) Binding to Heparin
J.Biol.Chem., 282, 2007
|
|
7CA5
 
 | | Cryo-EM structure of human GABA(B) receptor in apo state | | Descriptor: | Gamma-aminobutyric acid type B receptor subunit 1, Gamma-aminobutyric acid type B receptor subunit 2 | | Authors: | Kim, Y, Jeong, E, Jeong, J, Kim, Y, Cho, Y. | | Deposit date: | 2020-06-08 | | Release date: | 2020-11-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural Basis for Activation of the Heterodimeric GABA B Receptor.
J.Mol.Biol., 432, 2020
|
|
1GH6
 
 | | RETINOBLASTOMA POCKET COMPLEXED WITH SV40 LARGE T ANTIGEN | | Descriptor: | Large T antigen, Retinoblastoma-associated protein | | Authors: | Kim, H.Y, Cho, Y. | | Deposit date: | 2000-11-15 | | Release date: | 2001-11-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the inactivation of retinoblastoma tumor suppressor by SV40 large T antigen.
EMBO J., 20, 2001
|
|
4TUM
 
 | | Crystal structure of Ankyrin Repeat Domain of AKR2 | | Descriptor: | Ankyrin repeat domain-containing protein 2 | | Authors: | Gwon, G.H, Cho, Y. | | Deposit date: | 2014-06-24 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Ankyrin Repeat Domain of AKR2 Drives Chloroplast Targeting through Coincident Binding of Two Chloroplast Lipids.
Dev.Cell, 30, 2014
|
|
3A1J
 
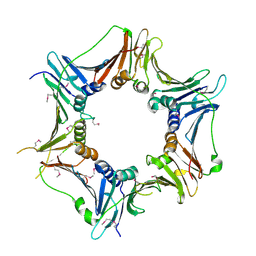 | | Crystal structure of the human Rad9-Hus1-Rad1 complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Checkpoint protein HUS1, ... | | Authors: | Sohn, S.Y, Cho, Y. | | Deposit date: | 2009-04-08 | | Release date: | 2009-06-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Human Rad9-Hus1-Rad1 Clamp
J.Mol.Biol., 390, 2009
|
|
3AV0
 
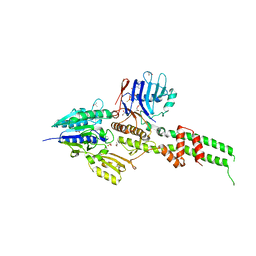 | | Crystal structure of Mre11-Rad50 bound to ATP S | | Descriptor: | DNA double-strand break repair protein mre11, DNA double-strand break repair rad50 ATPase, GLYCEROL, ... | | Authors: | Lim, H.S, Kim, J.S, Cho, Y. | | Deposit date: | 2011-02-18 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of the Mre11-Rad50-ATP S Complex: Understanding the Interplay between Mre11 and Rad50
To be Published
|
|
3AUY
 
 | | Crystal structure of Rad50 bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA double-strand break repair rad50 ATPase, MAGNESIUM ION | | Authors: | Lim, H.S, Cho, Y. | | Deposit date: | 2011-02-18 | | Release date: | 2011-05-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Mre11-Rad50-ATP S Complex: Understanding the Interplay between Mre11 and Rad50
To be Published
|
|
3AUZ
 
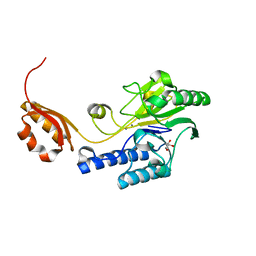 | | Crystal structure of Mre11 with manganese | | Descriptor: | DNA double-strand break repair protein mre11, GLYCEROL, MANGANESE (II) ION | | Authors: | Park, Y.B, Cho, Y. | | Deposit date: | 2011-02-18 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.206 Å) | | Cite: | Crystal Structure of the Mre11-Rad50-ATP S Complex: Understanding the Interplay between Mre11 and Rad50
To be Published
|
|
3AUX
 
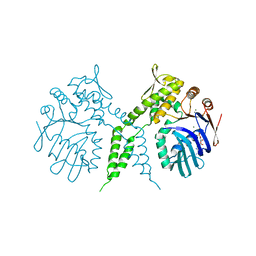 | | Crystal structure of Rad50 bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA double-strand break repair rad50 ATPase, MAGNESIUM ION | | Authors: | Lim, H.S, Cho, Y. | | Deposit date: | 2011-02-17 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Mre11-Rad50-ATP S Complex:Understanding the Interplay between Mre11 and Rad50
To be Published
|
|
7YQ3
 
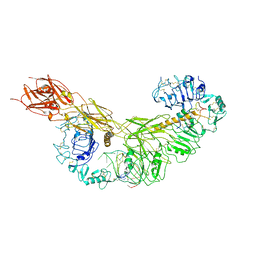 | | human insulin receptor bound with A43 DNA aptamer and insulin | | Descriptor: | IR-A43 aptamer, Insulin A chain, Insulin, ... | | Authors: | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
7YQ4
 
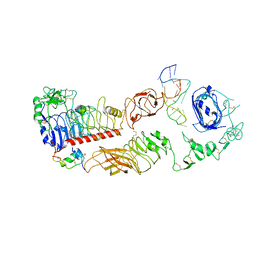 | | human insulin receptor bound with A62 DNA aptamer and insulin - locally refined | | Descriptor: | IR-A62 aptamer, Insulin A chain, Insulin, ... | | Authors: | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
7YQ6
 
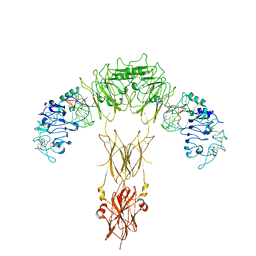 | | human insulin receptor bound with A62 DNA aptamer | | Descriptor: | IR-A62 aptamer, Isoform Short of Insulin receptor | | Authors: | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-09 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
7YQ5
 
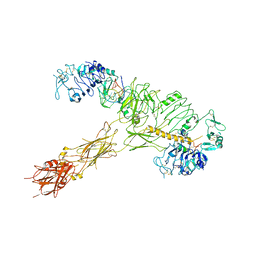 | | human insulin receptor bound with A62 DNA aptamer and insulin | | Descriptor: | IR-A62 aptamer, Insulin A chain, Insulin, ... | | Authors: | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-09 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
7WRS
 
 | |
7WRU
 
 | |
4R3Z
 
 | | Crystal structure of human ArgRS-GlnRS-AIMP1 complex | | Descriptor: | Aminoacyl tRNA synthase complex-interacting multifunctional protein 1, Arginine--tRNA ligase, cytoplasmic, ... | | Authors: | Fu, Y, Kim, Y, Cho, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4.033 Å) | | Cite: | Structure of the ArgRS-GlnRS-AIMP1 complex and its implications for mammalian translation
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1R6M
 
 | | Crystal Structure Of The tRNA Processing Enzyme Rnase pH From Pseudomonas Aeruginosa In Complex With Phosphate | | Descriptor: | PHOSPHATE ION, Ribonuclease PH | | Authors: | Choi, J.M, Park, E.Y, Kim, J.H, Chang, S.K, Cho, Y. | | Deposit date: | 2003-10-15 | | Release date: | 2004-02-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the functional importance of the hexameric ring structure of RNase PH
J.BIOL.CHEM., 279, 2004
|
|
4P0R
 
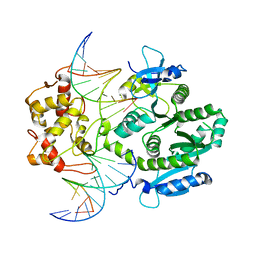 | | human Mus81-Eme1-3'flap DNA complex | | Descriptor: | Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, DNA ACGTGCTTACACACAGAGGTTAGGGTGAACTT, ... | | Authors: | Gwon, G.H, Baek, K, Cho, Y. | | Deposit date: | 2014-02-22 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (6.501 Å) | | Cite: | Crystal structures of the structure-selective nuclease Mus81-Eme1 bound to flap DNA substrates.
Embo J., 33, 2014
|
|
1R6L
 
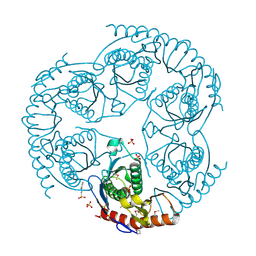 | | Crystal Structure Of The tRNA Processing Enzyme Rnase pH From Pseudomonas Aeruginosa | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Ribonuclease PH, SULFATE ION | | Authors: | Choi, J.M, Park, E.Y, Kim, J.H, Chang, S.K, Cho, Y. | | Deposit date: | 2003-10-15 | | Release date: | 2004-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the functional importance of the hexameric ring structure of RNase PH
J.BIOL.CHEM., 279, 2004
|
|
7YXA
 
 | | XFEL crystal structure of the human sphingosine 1 phosphate receptor 5 in complex with ONO-5430608 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[6-(2-naphthalen-1-ylethoxy)-2,3,4,5-tetrahydro-1H-3-benzazepin-3-ium-3-yl]butanoic acid, ... | | Authors: | Lyapina, E, Marin, E, Gusach, A, Orekhov, P, Gerasimov, A, Luginina, A, Vakhrameev, D, Ergasheva, M, Kovaleva, M, Khusainov, G, Khorn, P, Shevtsov, M, Kovalev, K, Okhrimenko, I, Bukhdruker, S, Popov, P, Hu, H, Weierstall, U, Liu, W, Cho, Y, Gushchin, I, Rogachev, A, Bourenkov, G, Park, S, Park, G, Huyn, H.J, Park, J, Gordeliy, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for receptor selectivity and inverse agonism in S1P 5 receptors.
Nat Commun, 13, 2022
|
|
4TUG
 
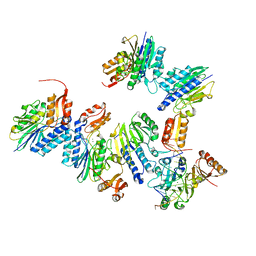 | | Crystal structure of MjMre11-DNA2 complex | | Descriptor: | DNA (5'-D(P*CP*TP*GP*TP*CP*CP*TP*AP*CP*GP*TP*GP*CP*CP*A)-3'), DNA (5'-D(P*GP*CP*AP*CP*GP*TP*AP*GP*GP*AP*CP*AP*GP*C)-3'), DNA double-strand break repair protein Mre11, ... | | Authors: | Sung, S, Cho, Y. | | Deposit date: | 2014-06-24 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | DNA end recognition by the Mre11 nuclease dimer: insights into resection and repair of damaged DNA.
Embo J., 33, 2014
|
|
4TUI
 
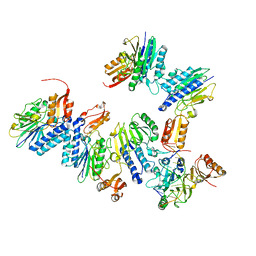 | | Crystal structure of MjMre11-DNA1 complex | | Descriptor: | DNA (5'-D(P*TP*CP*CP*TP*AP*CP*GP*TP*GP*CP*CP*AP*G)-3'), DNA (5'-D(P*TP*GP*GP*CP*AP*CP*GP*TP*AP*GP*GP*AP*C)-3'), DNA double-strand break repair protein Mre11 | | Authors: | Sung, S, Cho, Y. | | Deposit date: | 2014-06-24 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | DNA end recognition by the Mre11 nuclease dimer: insights into resection and repair of damaged DNA.
Embo J., 33, 2014
|
|
2KLO
 
 | | Structure of the Cdt1 C-terminal domain | | Descriptor: | DNA replication factor Cdt1 | | Authors: | Khayrutdinov, B.I, Bae, W.J, Yun, Y.M, Tsuyama, T, Kim, J.J, Hwang, E, Ryu, K.-S, Cheong, H.-K, Cheong, C, Karplus, P.A, Guntert, P, Tada, S, Jeon, Y.H, Cho, Y. | | Deposit date: | 2009-07-06 | | Release date: | 2009-10-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the Cdt1 C-terminal domain: Conservation of the winged helix fold in replication licensing factors
Protein Sci., 18, 2009
|
|
2MJP
 
 | | STRUCTURE-BASED IDENTIFICATION OF THE BIOCHEMICAL FUNCTION OF A HYPOTHETICAL PROTEIN FROM METHANOCOCCUS JANNASCHII:MJ0226 | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PYROPHOSPHATASE | | Authors: | Hwang, K.Y, Chung, J.H, Han, Y.S, Kim, S.H, Cho, Y, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 1999-01-27 | | Release date: | 2000-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based identification of a novel NTPase from Methanococcus jannaschii.
Nat.Struct.Biol., 6, 1999
|
|
