7ZNG
 
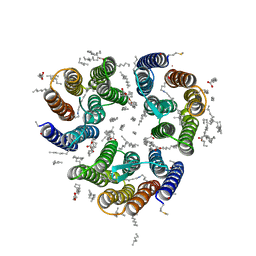 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the ground state at pH 8.2 at room temperature, 500-mks-long snapshots | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZNH
 
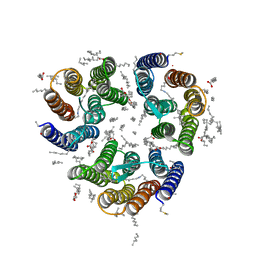 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the activated state at pH 8.2 at room temperature, 250-750-mks-snapshot | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZNI
 
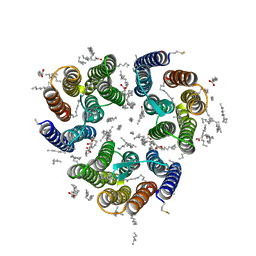 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the activated state at pH 8.2 at room temperature, 7.5-15-ms-snapshot | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6SJD
 
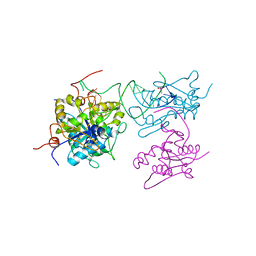 | | ZC3H12B-ribonuclease domain bound to RNA | | Descriptor: | CACODYLATE ION, MAGNESIUM ION, Probable ribonuclease ZC3H12B, ... | | Authors: | Morgunova, E, Bourenkov, G, Taipale, J. | | Deposit date: | 2019-08-13 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Binding specificities of human RNA-binding proteins toward structured and linear RNA sequences.
Genome Res., 30, 2020
|
|
6ZPL
 
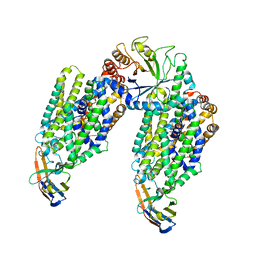 | | Inward-open structure of human glycine transporter 1 in complex with a benzoylisoindoline inhibitor, sybody Sb_GlyT1#7 and bound Na and Cl ions. | | Descriptor: | CHLORIDE ION, Endoglucanase H, SODIUM ION, ... | | Authors: | Shahsavar, A, Stohler, P, Bourenkov, G, Zimmermann, I, Siegrist, M, Guba, W, Pinard, E, Sinning, S, Seeger, M.A, Schneider, T.R, Dawson, R.J.P, Nissen, P. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.945 Å) | | Cite: | Structural insights into the inhibition of glycine reuptake.
Nature, 591, 2021
|
|
7QUO
 
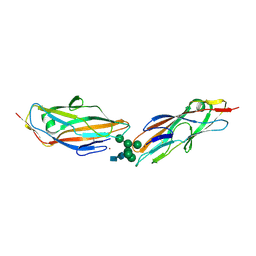 | | FimH lectin domain in complex with oligomannose-6 | | Descriptor: | FimH, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Bouckaert, J, Bourenkov, G.P. | | Deposit date: | 2022-01-18 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into a cooperative switch between one and two FimH bacterial adhesins binding pauci- and high-mannose type N-glycan receptors
J.Biol.Chem., 299, 2023
|
|
7NEV
 
 | | Structure of the hemiacetal complex between the SARS-CoV-2 Main Protease and Leupeptin | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H.M, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashhour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Xavier, P.L, Ullah, N, Andaleeb, H, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Zaitsev-Doyle, J.J, Rogers, C, Gieseler, H, Melo, D, Monteiro, D.C.F, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schluenzen, F, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Sun, X, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
8BY3
 
 | | FimH lectin domain in complex with oligomannose-6 | | Descriptor: | NICKEL (II) ION, SULFATE ION, Type 1 fimbrin D-mannose specific adhesin, ... | | Authors: | Bouckaert, J, Bourenkov, G.P. | | Deposit date: | 2022-12-11 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.186 Å) | | Cite: | Structural insights into a cooperative switch between one and two FimH bacterial adhesins binding pauci- and high-mannose type N-glycan receptors.
J.Biol.Chem., 299, 2023
|
|
5IJI
 
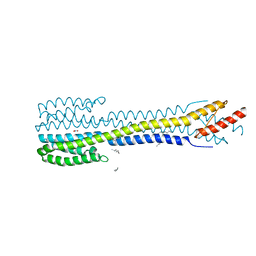 | | Fragment of nitrate/nitrite sensor histidine kinase NarQ (WT) in symmetric holo state | | Descriptor: | EICOSANE, NITRATE ION, Nitrate/nitrite sensor histidine kinase NarQ | | Authors: | Gushchin, I, Melnikov, I, Polovinkin, V, Ishchenko, A, Popov, A, Gordeliy, V. | | Deposit date: | 2016-03-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism of transmembrane signaling by sensor histidine kinases.
Science, 356, 2017
|
|
7BHD
 
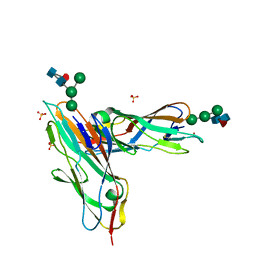 | | FimH in complex with alpha1,6 core-fucosylated oligomannose-3, crystallized in the trigonal space group | | Descriptor: | NICKEL (II) ION, SULFATE ION, Type 1 fimbrin D-mannose specific adhesin, ... | | Authors: | Bridot, C, Bouckaert, J, Krammer, E.-M. | | Deposit date: | 2021-01-11 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into a cooperative switch between one and two FimH bacterial adhesins binding pauci- and high-mannose type N-glycan receptors.
J.Biol.Chem., 299, 2023
|
|
7ABU
 
 | | Structure of SARS-CoV-2 Main Protease bound to RS102895 | | Descriptor: | 1'-[2-[4-(trifluoromethyl)phenyl]ethyl]spiro[1~{H}-3,1-benzoxazine-4,4'-piperidine]-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-08 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
2NAP
 
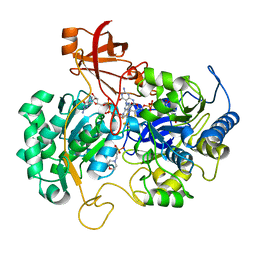 | | DISSIMILATORY NITRATE REDUCTASE (NAP) FROM DESULFOVIBRIO DESULFURICANS | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Dias, J.M, Than, M, Humm, A, Huber, R, Bourenkov, G, Bartunik, H, Bursakov, S, Calvete, J, Caldeira, J, Carneiro, C, Moura, J, Moura, I, Romao, M.J. | | Deposit date: | 1998-09-18 | | Release date: | 1999-09-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the first dissimilatory nitrate reductase at 1.9 A solved by MAD methods.
Structure Fold.Des., 7, 1999
|
|
5TJD
 
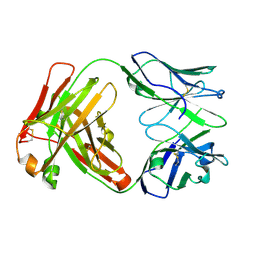 | | Computer-based rational design of improved functionality for antibody catalysts toward organophosphorus compounds | | Descriptor: | FAB A.17 L47K mutant HEAVY CHAIN, FAB A.17 L47K mutant Light Chain | | Authors: | Chatziefthimiou, S, Stepanova, A, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2016-10-04 | | Release date: | 2018-05-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 2020
|
|
1QLM
 
 | |
6SUI
 
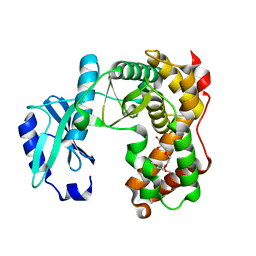 | | AMICOUMACIN KINASE AMIN | | Descriptor: | PENTAETHYLENE GLYCOL, Phosphotransferase enzyme family protein | | Authors: | Bourenkov, G.P, Mokrushina, Y.A, Terekhov, S.S, Smirnov, I.V, Gabibov, A.G, Altman, S. | | Deposit date: | 2019-09-14 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A kinase bioscavenger provides antibiotic resistance by extremely tight substrate binding.
Sci Adv, 6, 2020
|
|
6SUN
 
 | | Amicoumacin kinase hAmiN in complex with AMP-PNP, Ca2+ and Ami | | Descriptor: | APH domain-containing protein, amicoumacin kinase, Amicoumacin A, ... | | Authors: | Bourenkov, G.P, Mokrushina, Y.A, Terekhov, S.S, Smirnov, I.V, Gabibov, A.G, Altman, S. | | Deposit date: | 2019-09-16 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A kinase bioscavenger provides antibiotic resistance by extremely tight substrate binding.
Sci Adv, 6, 2020
|
|
6SV5
 
 | | Amicoumacin kinase AmiN in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Phosphotransferase enzyme family protein, amicoumacin kinase | | Authors: | Bourenkov, G.P, Mokrushina, Y.A, Terekhov, S.S, Smirnov, I.V, Gabibov, A.G, Altman, S. | | Deposit date: | 2019-09-17 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A kinase bioscavenger provides antibiotic resistance by extremely tight substrate binding.
Sci Adv, 6, 2020
|
|
6SUM
 
 | | Amicoumacin kinase hAmiN in complex with AMP-PNP, MG2+ and Ami | | Descriptor: | ACETATE ION, AMICOUMACIN KINASE, Amicoumacin A, ... | | Authors: | Bourenkov, G.P, Mokrushina, Y.A, Terekhov, S.S, Smirnov, I.V, Gabibov, A.G, Altman, S. | | Deposit date: | 2019-09-16 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A kinase bioscavenger provides antibiotic resistance by extremely tight substrate binding.
Sci Adv, 6, 2020
|
|
6SUL
 
 | | Amicoumacin kinase AmiN in complex with AMP-PNP, Mg2+ and Ami | | Descriptor: | Amicoumacin A, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bourenkov, G.P, Mokrushina, Y.A, Terekhov, S.S, Smirnov, I.V, Gabibov, A.G, Altman, S. | | Deposit date: | 2019-09-15 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A kinase bioscavenger provides antibiotic resistance by extremely tight substrate binding.
Sci Adv, 6, 2020
|
|
6YVK
 
 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 0.71 MGy exposure | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-04-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6YVL
 
 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 1.42 MGy exposure | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-04-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6YVM
 
 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 2.13 MGy exposure | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-04-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6YVN
 
 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 2.84 MGy exposure | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-04-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6YWU
 
 | | Human OMPD-domain of UMPS (K314AcK) in complex with UMP at 1.1 Angstroms resolution | | Descriptor: | GLYCEROL, SULFATE ION, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-04-30 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6YWT
 
 | |
