3RSR
 
 | | Crystal Structure of 5-NITP Inhibition of Yeast Ribonucleotide Reductase | | Descriptor: | 1-{2-DEOXY-5-O-[(R)-HYDROXY{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}PHOSPHORYL]-BETA-D-ERYTHRO-PENTOFURANOSYL}-5-NITRO-1H-INDOLE, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large chain 1 | | Authors: | Wan, Q, Mohammed, F, Jha, S, Motea, E, Berdis, A, Dealwis, C.G. | | Deposit date: | 2011-05-02 | | Release date: | 2012-11-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evaluating the therapeutic potential of a non-natural nucleotide that inhibits human ribonucleotide reductase.
Mol.Cancer Ther., 11, 2012
|
|
6JZP
 
 | |
7EO6
 
 | | X-ray structure analysis of xylanase | | Descriptor: | Endo-1,4-beta-xylanase, IODIDE ION | | Authors: | Wan, Q, Yi, Y, Xu, S. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization and structural analysis of a thermophilic GH11 xylanase from compost metatranscriptome.
Appl.Microbiol.Biotechnol., 105, 2021
|
|
7EO3
 
 | | X-ray structure analysis of beita-1,3-glucanase | | Descriptor: | 1,3-beta-glucanase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION | | Authors: | Wan, Q, Feng, J, Xu, S. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.141 Å) | | Cite: | Identification and structural analysis of a thermophilic beta-1,3-glucanase from compost
Bioresour Bioprocess, 8, 2021
|
|
6VJT
 
 | |
7RPK
 
 | | Cryo-EM structure of murine Dispatched in complex with Sonic hedgehog | | Descriptor: | (2S)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexanoyloxy)propyl hexanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | Deposit date: | 2021-08-03 | | Release date: | 2021-10-27 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
7RPJ
 
 | | Cryo-EM structure of murine Dispatched NNN mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Protein dispatched homolog 1 | | Authors: | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | Deposit date: | 2021-08-03 | | Release date: | 2021-10-27 | | Last modified: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
7RPH
 
 | | Cryo-EM structure of murine Dispatched 'R' conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | Deposit date: | 2021-08-03 | | Release date: | 2021-10-27 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
7RPI
 
 | | Cryo-EM structure of murine Dispatched 'T' conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | Deposit date: | 2021-08-03 | | Release date: | 2021-10-27 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
7Y9J
 
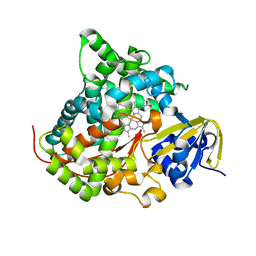 | | Crystal structure of P450 BM3-TMK from Bacillus megaterium in complex with 5-nitro-1,2-benzisoxazole | | Descriptor: | 5-nitro-1,2-benzoxazole, Bifunctional cytochrome P450/NADPH--P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, Q, Zhang, L.L, Liu, W.D, Huang, J.-W, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Engineering of a P450-based Kemp eliminase with a new mechanism
Chinese J Catal, 47, 2023
|
|
7Y9K
 
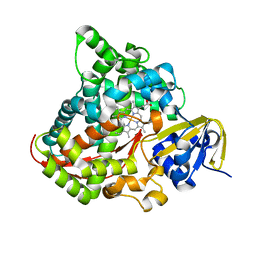 | | Crystal structure of P450 BM3-TMK from Bacillus megaterium | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, Q, Zhang, L.L, Liu, W.D, Huang, J.-W, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-25 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Engineering of a P450-based Kemp eliminase with a new mechanism
Chinese J Catal, 47, 2023
|
|
3RWT
 
 | |
3RWA
 
 | |
7DTE
 
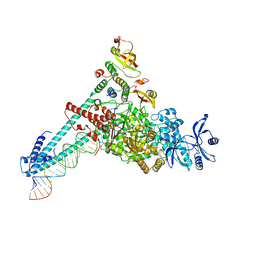 | | SARS-CoV-2 RdRP catalytic complex with T33-1 RNA | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA (33-MER), ... | | Authors: | Wang, Q, Gong, P. | | Deposit date: | 2021-01-04 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Remdesivir overcomes the S861 roadblock in SARS-CoV-2 polymerase elongation complex.
Cell Rep, 37, 2021
|
|
8H3D
 
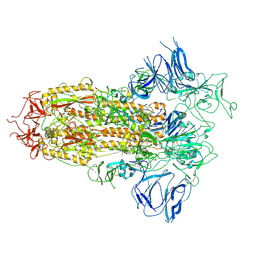 | | Structure of apo SARS-CoV-2 spike protein with one RBD up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Meng, F, Wang, Q, Xie, Y, Ni, X, Huang, N. | | Deposit date: | 2022-10-08 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | In Silico Discovery of Small Molecule Modulators Targeting the Achilles' Heel of SARS-CoV-2 Spike Protein.
Acs Cent.Sci., 9, 2023
|
|
8H3E
 
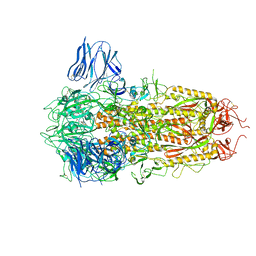 | | Complex structure of a small molecule (SPC-14) bound SARS-CoV-2 spike protein, closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-(6-nitro-2,3-dihydroindol-1-yl)-7-oxidanylidene-heptanoic acid, Spike glycoprotein,Fibritin | | Authors: | Meng, F, Wang, Q, Xie, Y, Ni, X, Huang, N. | | Deposit date: | 2022-10-08 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | In Silico Discovery of Small Molecule Modulators Targeting the Achilles' Heel of SARS-CoV-2 Spike Protein.
Acs Cent.Sci., 9, 2023
|
|
8HMH
 
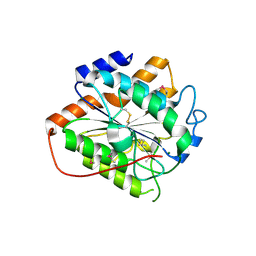 | | The closed state of RGLG2-VWA | | Descriptor: | E3 ubiquitin-protein ligase RGLG2, MAGNESIUM ION | | Authors: | Wang, Q. | | Deposit date: | 2022-12-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The regulation of RGLG2-VWA by Ca 2+ ions.
Biochim Biophys Acta Proteins Proteom, 1872, 2024
|
|
5DO2
 
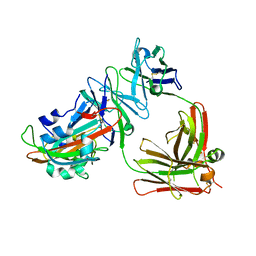 | | Complex structure of MERS-RBD bound with 4C2 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4C2 heavy chain, 4C2 light chain, ... | | Authors: | Li, Y, Wan, Y, Liu, P, Zhao, J, Lu, G, Qi, J, Wang, Q, Lu, X, Wu, Y, Liu, W, Yuen, K.Y, Perlman, S, Gao, G.F, Yan, J. | | Deposit date: | 2015-09-10 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.409 Å) | | Cite: | A humanized neutralizing antibody against MERS-CoV targeting the receptor-binding domain of the spike protein.
Cell Res., 25, 2015
|
|
6K9O
 
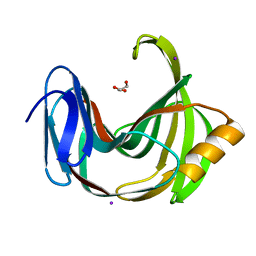 | | Crystal Structure Analysis of Protein | | Descriptor: | Endo-1,4-beta-xylanase 2, GLYCEROL, IODIDE ION | | Authors: | Li, C, Wan, Q. | | Deposit date: | 2019-06-17 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Studying the Role of a Single Mutation of a Family 11 Glycoside Hydrolase Using High-Resolution X-ray Crystallography.
Protein J., 39, 2020
|
|
6KWD
 
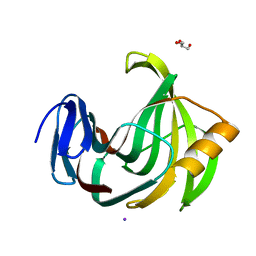 | | Crystal Structure Analysis of Endo-beta-1,4-Xylanase II Complexed with Xylotriose | | Descriptor: | Endo-1,4-beta-xylanase 2, GLYCEROL, IODIDE ION, ... | | Authors: | Li, C, Wan, Q. | | Deposit date: | 2019-09-06 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Studying the Role of a Single Mutation of a Family 11 Glycoside Hydrolase Using High-Resolution X-ray Crystallography.
Protein J., 39, 2020
|
|
6KW9
 
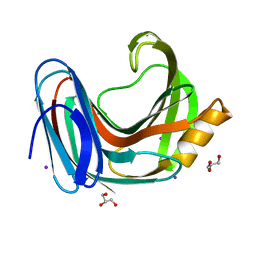 | |
3WP1
 
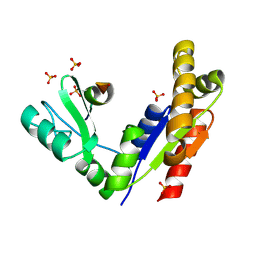 | | Phosphorylation-dependent interaction between tumor suppressors Dlg and Lgl | | Descriptor: | Disks large homolog 4, Lethal(2) giant larvae protein homolog 2, SULFATE ION | | Authors: | Zhu, J, Shang, Y, Wan, Q, Xia, Y, Chen, J, Du, Q, Zhang, M. | | Deposit date: | 2014-01-08 | | Release date: | 2014-03-19 | | Last modified: | 2014-04-30 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Phosphorylation-dependent interaction between tumor suppressors Dlg and Lgl
Cell Res., 24, 2014
|
|
3WP0
 
 | | Crystal structure of Dlg GK in complex with a phosphor-Lgl2 peptide | | Descriptor: | Disks large homolog 4, GLYCEROL, Lethal(2) giant larvae protein homolog 2 | | Authors: | Zhu, J, Shang, Y, Wan, Q, Xia, Y, Chen, J, Du, Q, Zhang, M. | | Deposit date: | 2014-01-08 | | Release date: | 2014-03-19 | | Last modified: | 2014-04-30 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | Phosphorylation-dependent interaction between tumor suppressors Dlg and Lgl
Cell Res., 24, 2014
|
|
3EL2
 
 | | Crystal Structure of Monomeric Actin Bound to Ca-ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, CALCIUM ION | | Authors: | Nair, U.B, Joel, P.B, Wan, Q, Lowey, S, Rould, M.A, Trybus, K.M. | | Deposit date: | 2008-09-19 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of monomeric actin bound to cytochalasin D.
J.Mol.Biol., 384, 2008
|
|
3EKS
 
 | | Crystal Structure of Monomeric Actin bound to Cytochalasin D | | Descriptor: | (3S,3aR,4S,6S,6aR,7E,10S,12R,13E,15R,15aR)-3-benzyl-6,12-dihydroxy-4,10,12-trimethyl-5-methylidene-1,11-dioxo-2,3,3a,4,5,6,6a,9,10,11,12,15-dodecahydro-1H-cycloundeca[d]isoindol-15-yl acetate, ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, ... | | Authors: | Nair, U.B, Joel, P.B, Wan, Q, Lowey, S, Rould, M.A, Trybus, K.M. | | Deposit date: | 2008-09-19 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of monomeric actin bound to cytochalasin D.
J.Mol.Biol., 384, 2008
|
|
