6NB3
 
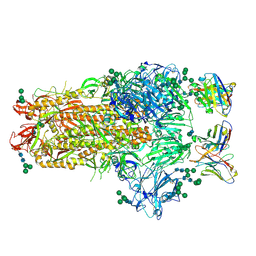 | | MERS-CoV complex with human neutralizing LCA60 antibody Fab fragment (state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LCA60 heavy chain, ... | | Authors: | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
2J6E
 
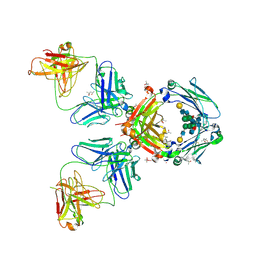 | | Crystal Structure of an Autoimmune Complex between a Human IgM Rheumatoid Factor and IgG1 Fc reveals a Novel Fc Epitope and Evidence for Affinity Maturation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CACODYLATE ION, ... | | Authors: | Duquerroy, S, Stura, E.A, Bressanelli, S, Browne, H, Beale, D, Hamon, M, Casali, P, Vaney, M.C, Rey, F.A, Sutton, B.J, Taussig, M.J. | | Deposit date: | 2006-09-28 | | Release date: | 2007-04-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a human autoimmune complex between IgM rheumatoid factor RF61 and IgG1 Fc reveals a novel epitope and evidence for affinity maturation.
J.Mol.Biol., 368, 2007
|
|
6NB4
 
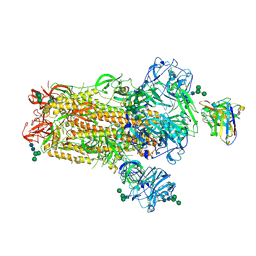 | | MERS-CoV S complex with human neutralizing LCA60 antibody Fab fragment (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LCA60 heavy chain, ... | | Authors: | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
6NB7
 
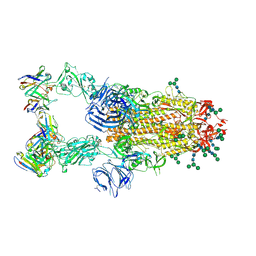 | | SARS-CoV complex with human neutralizing S230 antibody Fab fragment (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S230 heavy chain, ... | | Authors: | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
6NB6
 
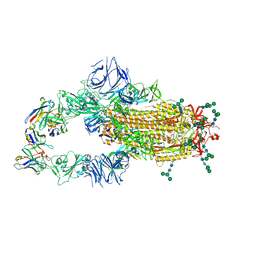 | | SARS-CoV complex with human neutralizing S230 antibody Fab fragment (state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S230 heavy chain, ... | | Authors: | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
6NZK
 
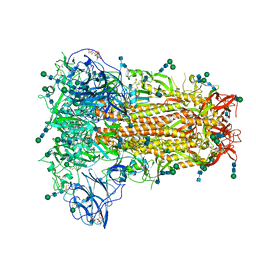 | | Structural basis for human coronavirus attachment to sialic acid receptors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike surface glycoprotein, ... | | Authors: | Tortorici, M.A, Walls, A.C, Lang, Y, Wang, C, Li, Z, Koerhuis, D, Boons, G.J, Bosch, B.J, Rey, F.A, de Groot, R, Veesler, D. | | Deposit date: | 2019-02-13 | | Release date: | 2019-06-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for human coronavirus attachment to sialic acid receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6NB5
 
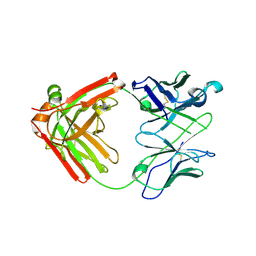 | | Crystal structure of anti- MERS-CoV human neutralizing LCA60 antibody Fab fragment | | Descriptor: | LCA60 antigen-binding (Fab) fragment, heavy chain, light chain | | Authors: | Walls, A.J, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, J, Quispe, J, Cameroni, E, Gopal, R, Dai, M, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
6NB8
 
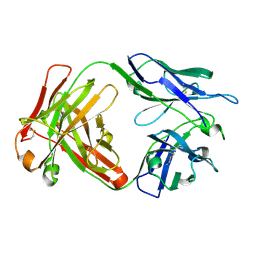 | | Crystal structure of anti- SARS-CoV human neutralizing S230 antibody Fab fragment | | Descriptor: | S230 antigen-binding (Fab) fragment, heavy chain, light chain | | Authors: | Walls, A.J, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, J, Quispe, J, Cameroni, E, Gopal, R, Dai, M, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
3UC0
 
 | | Crystal structure of domain I of the envelope glycoprotein ectodomain from dengue virus serotype 4 in complex with the fab fragment of the chimpanzee monoclonal antibody 5H2 | | Descriptor: | GLYCEROL, Heavy chain, monoclonal antibody 5H2, ... | | Authors: | Cockburn, J.J.B, Stura, E.A, Navarro-Sanchez, M.E, Rey, F.A. | | Deposit date: | 2011-10-25 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural insights into the neutralization mechanism of a higher primate antibody against dengue virus.
Embo J., 31, 2012
|
|
3UZQ
 
 | | Crystal structure of the dengue virus serotype 1 envelope protein domain III in complex with the variable domains of Mab 4E11 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Cockburn, J.J.B, Navarro Sanchez, M.E, Fretes, N, Urvoas, A, Staropoli, I, Kikuti, C.M, Coffey, L.L, Arenzana Seisdedos, F, Bedouelle, H, Rey, F.A. | | Deposit date: | 2011-12-07 | | Release date: | 2012-02-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanism of dengue virus broad cross-neutralization by a monoclonal antibody.
Structure, 20, 2012
|
|
5MF1
 
 | |
3UZV
 
 | | Crystal structure of the dengue virus serotype 2 envelope protein domain III in complex with the variable domains of Mab 4E11 | | Descriptor: | ETHANOL, anti-dengue Mab 4E11, envelope protein | | Authors: | Cockburn, J.J.B, Navarro Sanchez, M.E, Fretes, N, Urvoas, A, Staropoli, I, Kikuti, C.M, Coffey, L.L, Arenzana Seisdedos, F, Bedouelle, H, Rey, F.A. | | Deposit date: | 2011-12-07 | | Release date: | 2012-02-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of dengue virus broad cross-neutralization by a monoclonal antibody.
Structure, 20, 2012
|
|
3UYP
 
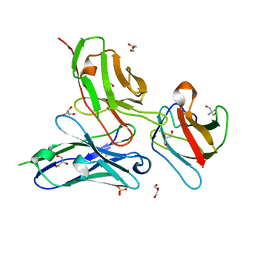 | | Crystal structure of the dengue virus serotype 4 envelope protein domain III in complex with the variable domains of Mab 4E11 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Envelope protein, GLYCEROL, ... | | Authors: | Cockburn, J.J.B, Navarro Sanchez, M.E, Fretes, N, Urvoas, A, Staropoli, I, Kikuti, C.M, Coffey, L.L, Arenzana Seisdedos, F, Bedouelle, H, Rey, F.A. | | Deposit date: | 2011-12-06 | | Release date: | 2012-02-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of dengue virus broad cross-neutralization by a monoclonal antibody.
Structure, 20, 2012
|
|
3UZE
 
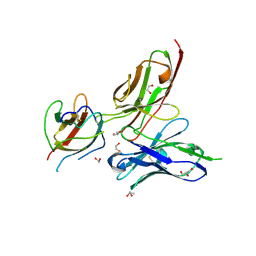 | | Crystal structure of the dengue virus serotype 3 envelope protein domain III in complex with the variable domains of Mab 4E11 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Envelope protein, ... | | Authors: | Cockburn, J.J.B, Navarro Sanchez, M.E, Fretes, N, Urvoas, A, Staropoli, I, Kikuti, C.M, Coffey, L.L, Arenzana Seisdedos, F, Bedouelle, H, Rey, F.A. | | Deposit date: | 2011-12-07 | | Release date: | 2012-02-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Mechanism of dengue virus broad cross-neutralization by a monoclonal antibody.
Structure, 20, 2012
|
|
3UAJ
 
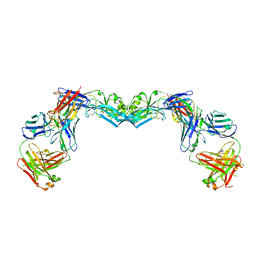 | | Crystal structure of the envelope glycoprotein ectodomain from dengue virus serotype 4 in complex with the fab fragment of the chimpanzee monoclonal antibody 5H2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, monoclonal antibody 5H2, ... | | Authors: | Cockburn, J.J.B, Stura, E.A, Navarro-Sanchez, M.E, Rey, F.A. | | Deposit date: | 2011-10-21 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.232 Å) | | Cite: | Structural insights into the neutralization mechanism of a higher primate antibody against dengue virus.
Embo J., 31, 2012
|
|
2VF1
 
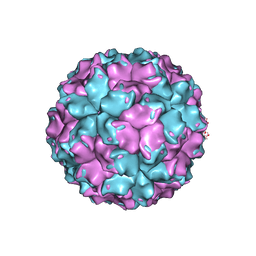 | | X-ray crystallographic structure of the picobirnavirus capsid | | Descriptor: | CAPSID PROTEIN | | Authors: | Duquerroy, S, Da Costa, B, Vigouroux, A, Lepault, J, Navaza, J, Delmas, B, Rey, F.A. | | Deposit date: | 2007-10-29 | | Release date: | 2008-12-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Picobirnavirus Crystal Structure Provides Functional Insights Into Virion Assembly and Cell Entry.
Embo J., 28, 2009
|
|
2XT1
 
 | | Crystal structure of the HIV-1 capsid protein C-terminal domain (146- 231) in complex with a camelid VHH. | | Descriptor: | CAMELID VHH 9, GAG POLYPROTEIN, GLYCEROL | | Authors: | Igonet, S, Vaney, M.C, Bartonova, V, Helma, J, Rothbauer, U, Leonhardt, H, Stura, E, Krausslich, H.-G, Rey, F.A. | | Deposit date: | 2010-10-03 | | Release date: | 2011-10-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Targeting HIV-1 Virion Formation with Nanobodies -Implications for the Design of Assembly Inhibitors
To be Published
|
|
2XXC
 
 | | Crystal structure of a camelid VHH raised against the HIV-1 capsid protein C-terminal domain. | | Descriptor: | CAMELID VHH 9 | | Authors: | Igonet, S, Vaney, M.C, Bartonova, V, Helma, J, Rothbauer, U, Leonhardt, H, Stura, E, Krausslich, H.-G, Rey, F.A. | | Deposit date: | 2010-11-10 | | Release date: | 2011-10-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Targeting HIV-1 Virion Formation with Nanobodies -Implications for the Design of Assembly Inhibitors
To be Published
|
|
2XHU
 
 | | HCV-J4 NS5B Polymerase Orthorhombic Crystal Form | | Descriptor: | RNA-directed RNA polymerase, SULFATE ION | | Authors: | Harrus, D, Ahmed-El-Sayed, N, Simister, P.C, Miller, S, Triconnet, M, Hagedorn, C.H, Mahias, K, Rey, F.A, Astier-Gin, T, Bressanelli, S. | | Deposit date: | 2010-06-21 | | Release date: | 2010-08-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Further Insights Into the Roles of GTP and the C- Terminus of the Hepatitis C Virus Polymerase in the Initiation of RNA Synthesis
J.Biol.Chem., 285, 2010
|
|
1WYY
 
 | | Post-fusion hairpin conformation of the sars coronavirus spike glycoprotein | | Descriptor: | CHLORIDE ION, E2 Glycoprotein | | Authors: | Duquerroy, S, Vigouroux, A, Rottier, P.J.M, Rey, F.A, Bosch, B.J. | | Deposit date: | 2005-02-18 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Central ions and lateral asparagine/glutamine zippers stabilize the post-fusion hairpin conformation of the SARS coronavirus spike glycoprotein
Virology, 335, 2005
|
|
2XI3
 
 | | HCV-H77 NS5B Polymerase Complexed With GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA-directed RNA polymerase | | Authors: | Harrus, D, Ahmed-El-Sayed, N, Simister, P.C, Miller, S, Triconnet, M, Hagedorn, C.H, Mahias, K, Rey, F.A, Astier-Gin, T, Bressanelli, S. | | Deposit date: | 2010-06-25 | | Release date: | 2010-08-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Further Insights Into the Roles of GTP and the C- Terminus of the Hepatitis C Virus Polymerase in the Initiation of RNA Synthesis
J.Biol.Chem., 285, 2010
|
|
2XI2
 
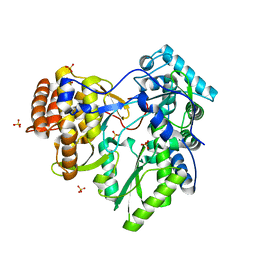 | | HCV-H77 NS5B Apo Polymerase | | Descriptor: | RNA-directed RNA polymerase, SULFATE ION | | Authors: | Harrus, D, Ahmed-El-Sayed, N, Simister, P.C, Miller, S, Triconnet, M, Hagedorn, C.H, Mahias, K, Rey, F.A, Astier-Gin, T, Bressanelli, S. | | Deposit date: | 2010-06-25 | | Release date: | 2010-08-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Further Insights Into the Roles of GTP and the C- Terminus of the Hepatitis C Virus Polymerase in the Initiation of RNA Synthesis
J.Biol.Chem., 285, 2010
|
|
2XXM
 
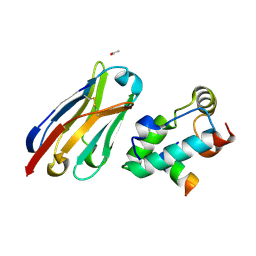 | | Crystal structure of the HIV-1 capsid protein C-terminal domain in complex with a camelid VHH and the CAI peptide. | | Descriptor: | ACETATE ION, CAMELID VHH 9, CAPSID PROTEIN P24, ... | | Authors: | Igonet, S, Vaney, M.C, Bartonova, V, Helma, J, Rothbauer, U, Leonhardt, H, Stura, E, Krausslich, H.-G, Rey, F.A. | | Deposit date: | 2010-11-10 | | Release date: | 2011-10-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Targeting HIV-1 Virion Formation with Nanobodies -Implications for the Design of Assembly Inhibitors
To be Published
|
|
2XHW
 
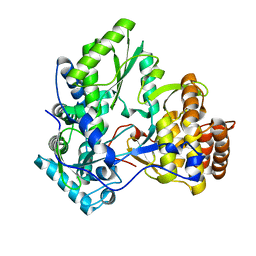 | | HCV-J4 NS5B Polymerase Trigonal Crystal Form | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Harrus, D, Ahmed-El-Sayed, N, Simister, P.C, Miller, S, Triconnet, M, Hagedorn, C.H, Mahias, K, Rey, F.A, Astier-Gin, T, Bressanelli, S. | | Deposit date: | 2010-06-21 | | Release date: | 2010-08-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Further Insights Into the Roles of GTP and the C- Terminus of the Hepatitis C Virus Polymerase in the Initiation of RNA Synthesis
J.Biol.Chem., 285, 2010
|
|
2XHV
 
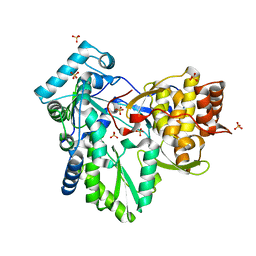 | | HCV-J4 NS5B Polymerase Point Mutant Orthorhombic Crystal Form | | Descriptor: | MAGNESIUM ION, RNA-directed RNA polymerase, SULFATE ION | | Authors: | Harrus, D, Ahmed-El-Sayed, N, Simister, P.C, Miller, S, Triconnet, M, Hagedorn, C.H, Mahias, K, Rey, F.A, Astier-Gin, T, Bressanelli, S. | | Deposit date: | 2010-06-21 | | Release date: | 2010-08-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Further Insights Into the Roles of GTP and the C- Terminus of the Hepatitis C Virus Polymerase in the Initiation of RNA Synthesis
J.Biol.Chem., 285, 2010
|
|
