4JHZ
 
 | | Structure of E. coli beta-Glucuronidase bound with a novel, potent inhibitor 2-[4-(1,3-benzodioxol-5-ylmethyl)piperazin-1-yl]-N-[(1S,2S,5S)-2,5-dimethoxycyclohexyl]acetamide | | Descriptor: | 2-[4-(1,3-benzodioxol-5-ylmethyl)piperazin-1-yl]-N-[(1S,2S,5S)-2,5-dimethoxycyclohexyl]acetamide, Beta-glucuronidase | | Authors: | Roberts, A.B, Wallace, B.D, Redinbo, M.R. | | Deposit date: | 2013-03-05 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.831 Å) | | Cite: | Molecular Insights into Microbial beta-Glucuronidase Inhibition to Abrogate CPT-11 Toxicity.
Mol.Pharmacol., 84, 2013
|
|
4JKL
 
 | |
4J5W
 
 | | Crystal Structure of the apo-PXR/RXRalpha LBD Heterotetramer Complex | | Descriptor: | MAGNESIUM ION, Nuclear receptor subfamily 1 group I member 2, Nuclear receptor coactivator 1, ... | | Authors: | Wallace, B.D, Betts, L, Redinbo, M.R. | | Deposit date: | 2013-02-10 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Functional Analysis of the Human Nuclear Xenobiotic Receptor PXR in Complex with RXRalpha.
J.Mol.Biol., 425, 2013
|
|
1ILH
 
 | |
1ILG
 
 | |
1K4Y
 
 | | Crystal Structure of Rabbit Liver Carboxylesterase in Complex with 4-piperidino-piperidine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-PIPERIDINO-PIPERIDINE, LIVER CARBOXYLESTERASE, ... | | Authors: | Bencharit, S, Morton, C.L, Howard-Williams, E.L, Danks, M.K, Potter, P.M, Redinbo, M.R. | | Deposit date: | 2001-10-09 | | Release date: | 2002-05-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into CPT-11 activation by mammalian carboxylesterases.
Nat.Struct.Biol., 9, 2002
|
|
1LPQ
 
 | | Human DNA Topoisomerase I (70 Kda) In Non-Covalent Complex With A 22 Base Pair DNA Duplex Containing an 8-oxoG Lesion | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*TP*(8OG)P*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', 5'-D(*AP*AP*AP*AP*AP*TP*TP*TP*TP*TP*CP*CP*AP*AP*GP*TP*CP*TP*TP*TP*TP*T)-3', DNA topoisomerase I | | Authors: | Lesher, D.T, Pommier, Y, Stewart, L, Redinbo, M.R. | | Deposit date: | 2002-05-08 | | Release date: | 2002-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | 8-Oxoguanine rearranges the active site of human topoisomerase I
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1M13
 
 | | Crystal Structure of the Human Pregane X Receptor Ligand Binding Domain in Complex with Hyperforin, a Constituent of St. John's Wort | | Descriptor: | 4-HYDROXY-5-ISOBUTYRYL-6-METHYL-1,3,7-TRIS-(3-METHYL-BUT-2-ENYL)-6-(4-METHYL-PENT-3-ENYL)-BICYCLO[3.3.1]NON-3-ENE-2,9-DIONE, Orphan Nuclear Receptor PXR | | Authors: | Watkins, R.E, Maglich, J.M, Moore, L.B, Wisely, G.B, Noble, S.M, Davis-Searles, P.R, Lambert, M.H, Kliewer, S.A, Redinbo, M.R. | | Deposit date: | 2002-06-17 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.1 A Crystal Structure of Human PXR in Complex with the St. John's Wort Compound Hyperforin
Biochemistry, 42, 2003
|
|
1MX9
 
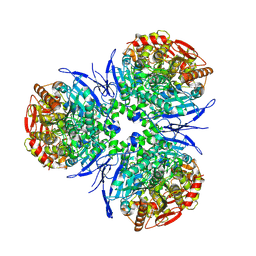 | | Crystal Structure of Human Liver Carboxylesterase in complexed with naloxone methiodide, a heroin analogue | | Descriptor: | (5A,17R)-4,5-EPOXY-3,14-DIHYDROXY-17-METHYL-6-OXO-17-(2-PROPENYL)-MORPHINANIUM, 2-acetamido-2-deoxy-beta-D-glucopyranose, liver Carboxylesterase I | | Authors: | Bencharit, S, Morton, C.L, Xue, Y, Potter, P.M, Redinbo, M.R. | | Deposit date: | 2002-10-01 | | Release date: | 2003-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis of Heroin and Cocaine Metabolism by a Promiscuous Human Drug-Processing Enzyme
Nat.Struct.Biol., 10, 2003
|
|
1MX5
 
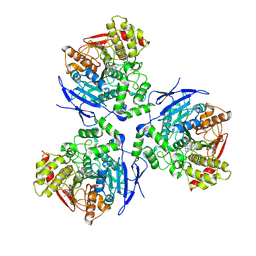 | | Crystal Structure of Human Liver Carboxylesterase in complexed with homatropine, a cocaine analogue | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, HOMOTROPINE, ... | | Authors: | Bencharit, S, Morton, C.L, Xue, Y, Potter, P.M, Redinbo, M.R. | | Deposit date: | 2002-10-01 | | Release date: | 2003-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Heroin and Cocaine Metabolism by a Promiscuous Human Drug-Processing Enzyme
Nat.Struct.Biol., 10, 2003
|
|
1MX1
 
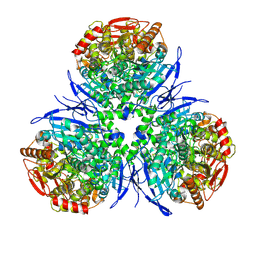 | | Crystal Structure of Human Liver Carboxylesterase in complex with tacrine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Bencharit, S, Morton, C.L, Hyatt, J.L, Kuhn, P, Danks, M.K, Potter, P.M, Redinbo, M.R. | | Deposit date: | 2002-10-01 | | Release date: | 2003-04-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Human Carboxylesterase 1 Complexed with the Alzheimer's
Drug Tacrine: From Binding Promiscuity to Selective Inhibition
CHEM.BIOL., 10, 2003
|
|
8DTQ
 
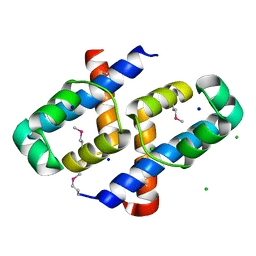 | | Crystal Structure of Staphylococcus aureus pSK41 Cop | | Descriptor: | CHLORIDE ION, Helix-turn-helix domain, SODIUM ION | | Authors: | Walton, W.G, Eakes, T.C, Redinbo, M.R, McLaughlin, K.J. | | Deposit date: | 2022-07-26 | | Release date: | 2023-08-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | pSK41/pGO1-family conjugative plasmids of Staphylococcus aureus encode a cryptic repressor of replication.
Plasmid, 128, 2023
|
|
3GS3
 
 | |
3K4D
 
 | | Crystal structure of E. coli beta-glucuronidase with the glucaro-d-lactam inhibitor bound | | Descriptor: | (2S,3R,4S,5R)-3,4,5-trihydroxy-6-oxopiperidine-2-carboxylic acid, Beta-glucuronidase | | Authors: | Wallace, B.D, Redinbo, M.R. | | Deposit date: | 2009-10-05 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Alleviating cancer drug toxicity by inhibiting a bacterial enzyme.
Science, 330, 2010
|
|
3K9B
 
 | |
3K4A
 
 | |
3K46
 
 | |
3LPF
 
 | | Structure of E. coli beta-Glucuronidase bound with a novel, potent inhibitor 1-((6,7-dimethyl-2-oxo-1,2-dihydroquinolin-3-yl)methyl)-1-(2-hydroxyethyl)-3-(3-methoxyphenyl)thiourea | | Descriptor: | 1-[(6,7-dimethyl-2-oxo-1,2-dihydroquinolin-3-yl)methyl]-1-(2-hydroxyethyl)-3-(3-methoxyphenyl)thiourea, Beta-glucuronidase | | Authors: | Wallace, B.D, Redinbo, M.R. | | Deposit date: | 2010-02-05 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Alleviating cancer drug toxicity by inhibiting a bacterial enzyme.
Science, 330, 2010
|
|
3LPG
 
 | |
3MBO
 
 | | Crystal Structure of the Glycosyltransferase BaBshA bound with UDP and L-malate | | Descriptor: | D-MALATE, GLYCEROL, Glycosyltransferase, ... | | Authors: | Wallace, B.D, Claiborne, A, Redinbo, M.R. | | Deposit date: | 2010-03-25 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.307 Å) | | Cite: | Characterization of the N-Acetyl-alpha-d-glucosaminyl
l-Malate Synthase and Deacetylase Functions for Bacillithiol Biosynthesis
in Bacillus anthracis
Biochemistry, 49, 2010
|
|
6D7J
 
 | |
7KFO
 
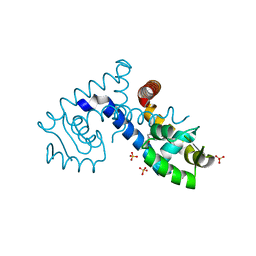 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to Indole 3 acetic acid | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-14 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7KGZ
 
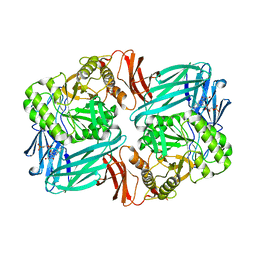 | |
7KFQ
 
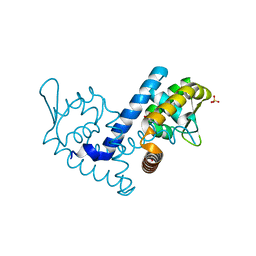 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to 1H-Indole-3-Carboxylic Acid | | Descriptor: | 1H-INDOLE-3-CARBOXYLIC ACID, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-14 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7KH3
 
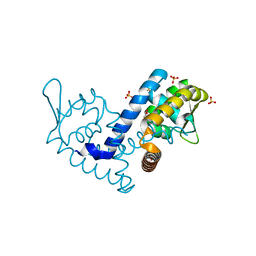 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to Indole 3 propionic acid | | Descriptor: | INDOLYLPROPIONIC ACID, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-19 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
